Isolation of Restriction Fragments from Agarose Gels by Collection onto DEAE Cellulose
PrincipleA DNA restriction fragment is isolated by collection on DEAE cellulose paper during electrophoresis, then washed from the DEAE with a high salt buffer, cleaned, precipitated, and resuspended in a small volume. Recovery of 50%–90% of the bound DNA can be expected; however, fragments larger than 7 kb have lower yields. DNA prepared this way is suitable for subcloning.
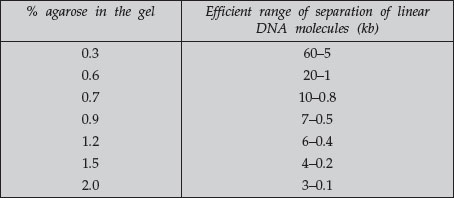
A DNA fragment of a given size migrates at different rates through gels containing different concentrations of agarose. By using a gel at the appropriate gel concentration, it is possible to resolve well the DNA of interest. Use the following table as a guide for determining the agarose concentration to use.
 |
|---|
Time Required
- 3–4 hours on Day 1
- 2–4 hours on Day 2
- Schleicher & Schuell NA-45 DEAE membrane.
Day 1
Isolating the fragment:
- Run the restriction digest and the appropriate size markers on a 1X Tris- Borate agarose gel with ethidium bromide. Be sure to leave at least 1–2 wells between samples. Run the gel until the DNA bands are well separated (visualize on the long-wavelength UV lightbox).
- Cut a slit just ahead of the band of interest using a sharp sterile razor blade or scalpel. Using blunt-edged forceps (such as Millipore forceps), carefully insert an NA-45 paper into the slit (prewet and cut NA-45 to the width of the band, see preparation of the NA-45 below).
- Place the gel in fresh 1X Tris-Borate buffer, and run the gel until the fragment has moved out of the gel and stopped by the NA-45. Monitor the progress of the band with the handheld long-wavelength UV light. Do not allow other bands of higher molecular weight to run onto the NA-45.
- Remove the NA-45 paper, rinse in NET buffer, and place in a labeled eppendorf tube. Add sufficient high-salt NET buffer to cover most of the membrane (typically 150–300 µL). Spin 5 seconds in a microcentrifuge to submerge the entire strip. Place at 65°C for 1 hour, mixing frequently, and respinning if the membrane rides up the side of the tube.
- Transfer the buffer (+DNA fragment) to a clean, labeled tube. Wash the membrane (in the original tube) with 50 mL high-salt NET buffer and add the wash to the DNA fragment tube.
- To remove ethidium bromide, extract twice with 3 volumes water-saturated n-butanol.
- Precipitate the DNA with 2.5 volumes of ethanol at –20°C for at least 1 hour (can sit overnight in the freezer).
- Pellet the DNA (for 20 minutes at high speed in a microcentrifuge) and resuspend in 50 µL TE. Reprecipitate with sodium acetate to remove any residual NaCl. Add 5 mL 3M Na-acetate, and 120 µL ethanol, hold at –20°C for 2 hours or more, pellet as before, and resuspend in an appropriate amount of TE.
Solutions
- Preparation of the DEAE cellulose membrane. Schleicher and Schuell NA-45 can be used as supplied by the manufacturer, with prewetting in sterile dH2O. However, the binding capacity of the membrane is increased with the following: a 10-minute soak in 10 mM EDTA pH 7.6, then 5 minutes in 0.5 N NaOH, followed by several rapid washes in sterile dH2O. Membranes can be stored for several weeks in sterile dH2O at 4°C.
- NET buffer (500 mL). 25 mL 3 M NaCl, 150 mM NaCl, 100 mL 0.5 M EDTA, 100 mM EDTA,10 mL 1 M Tris pH 7.5, 20 mM Tris pH 7.5, 365 mL dH2O. Autoclave to sterilize.
- High salt NET buffer (500 mL). 166.7 mL 3M NaCl, 1 M NaCl, 100 mL 0.5 M EDTA, 100 mM EDTA,10 mL 1M Tris pH 7.5, 20 mM Tris pH7.5, 223.3 mL dH2O. Autoclave to sterilize.




