PCR Methods (Polymerase Chain Reaction)
Who would have thought a bacterium hanging out in a hot spring in Yellowstone National Park would spark a revolutionary new laboratory technique? The polymerase chain reaction, now widely used in research laboratories and doctors’ offices, relies on the ability of DNA-copying enzymes to remain stable at high temperatures. No problem for Thermus aquaticus, the sultry bacterium from Yellowstone that now helps scientists produce millions of copies of a single DNA segment in a matter of hours.In nature, most organisms copy their DNA in the same way. The PCR mimics this process, only it does it in a test tube. When any cell divides, enzymes called polymerases make a copy of all the DNA in each chromosome. The first step in this process is to “unzip” the 2 DNA chains of the double helix. As the 2 strands separate, DNA polymerase makes a copy, using each strand as a template.
The 4 nucleotide bases, the building blocks of every piece of DNA, are represented by the letters A, C, G, and T, which stand for their chemical names: adenine, cytosine, guanine, and thymine. The A on one strand always pairs with the T on the other, whereas C always pairs with G. The 2 strands are said to be complementary to each other.
To copy DNA, polymerase requires 2 other components: a supply of the 4 nucleotide bases and something called a primer. DNA polymerases, whether from humans, bacteria, or viruses, cannot copy a chain of DNA without a short
sequence of nucleotides to “prime” the process, or get it started. So the cell has another enzyme called a primase that actually makes the first few nucleotides of the copy. This stretch of DNA is called a primer. Once the primer is made, the polymerase can take over, making the rest of the new chain.
A PCR vial contains all the necessary components for DNA duplication: a piece of DNA, large quantities of the 4 nucleotides, large quantities of the primer sequence, and DNA polymerase. The polymerase is the Taq polymerase, named for Thermus aquaticus, from which it was isolated.
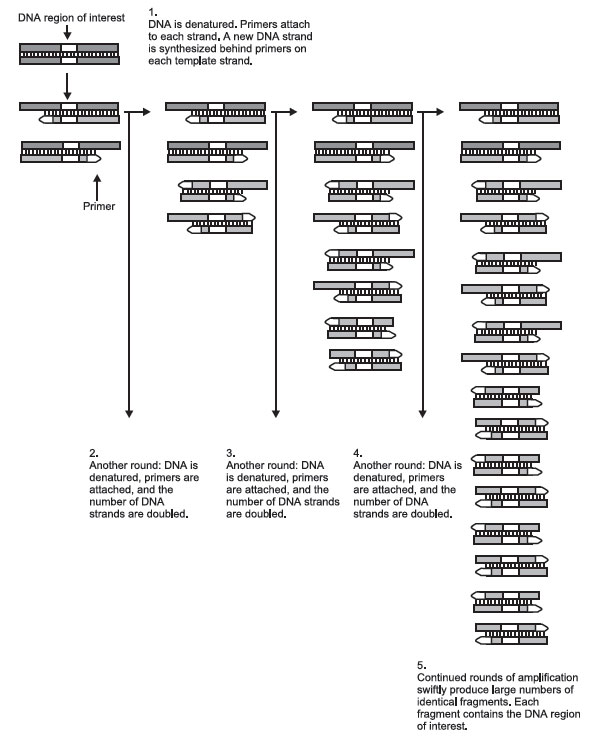
The 3 parts of the polymerase chain reaction are carried out in the same vial, but at different temperatures. The first part of the process separates the 2 DNA chains in the double helix. This is done simply by heating the vial to 90°C–95°C (about 165°F) for 30 seconds.
But the primers cannot bind to the DNA strands at such a high temperature, so the vial is cooled to 55°C (about 100°F). At this temperature, the primers bind or “anneal” to the ends of the DNA strands. This takes about 20 seconds.
The final step of the reaction is to make a complete copy of the templates. Since the Taq polymerase works best at around 75°C (the temperature of the hot springs where the bacterium was discovered), the temperature of the vial is raised.
The Taq polymerase begins adding nucleotides to the primer and eventually makes a complementary copy of the template. If the template contains an A nucleotide, the enzyme adds on a T nucleotide to the primer. If the template contains a G, it adds a C to the new chain, and so on, to the end of the DNA strand. This completes 1 PCR cycle.
The 3 steps in the polymerase chain reaction—the separation, of the strands, annealing the primer to the template, and the synthesis of new strands—take less than two minutes. Each is carried out in the same vial. At the end of a cycle, each piece of DNA in the vial has been duplicated.
But the cycle can be repeated 30 or more times. Each newly synthesized DNA piece can act as a new template, so after 30 cycles, 1 million copies of a single piece of DNA can be produced! Taking into account the time it takes to change the temperature of the reaction vial, 1 million copies can be ready in about 3 hours.
 |
|---|
In one application of the technology, small samples of DNA, such as those found in a strand of hair at a crime scene, can produce sufficient copies to carry out forensic tests.




