Protein Translocation into Mitochondria
I. INTRODUCTIONMitochondria from different sources such as rat liver, rabbit brain, or yeast can be isolated as intact organelles. Isolated mitochondria are able to respire, maintain a membrane potential across their inner membrane, possess an active ATP synthase, and shuttle nucleotides across their membranes. In addition, even a process as complicated as import of mitochondrial precursor proteins can be studied outside the living cell. For this purpose, radiolabeled precursor proteins, synthesized in an in vitro transcription/ translation system, are mixed with isolated mitochondria (Glick, 1991; Melton et al., 1984). In the presence of ATP, precursor proteins will cross the mitochondrial membranes, become processed to their mature form, and fold to their native state. Building on this basic "import assay," sophisticated experiments have been developed and the results of these experiments provide most of what we know about mitochondrial import today (Neupert, 1997; Pfanner and Geissler, 2001).
This article describes a standard protocol for the in vitro synthesis of a radiolabeled precursor protein and the subsequent import of this precursor into isolated yeast mitochondria. As an example, we have selected the precursor protein yeast malate dehydrogenase (Dubaqui6 et al., 1998). The N-terminal presequence of the yeast malate dehydrogenase precursor, as is of most mitochondrial precursor proteins, is removed by a protease localized in the mitochondrial matrix (Jensen and Yaffe, 1988). The mRNA of the precursor protein is transcribed with SP6 RNA polymerase (Melton et al., 1984).
II. MATERIALS AND INSTRUMENTATION
SP6 RNA polymerase (Cat. No. 810 274); RNase inhibitor from human placenta (Cat. No. 799 017); set of ATP, CTP, GTP, UTP, lithium salts, 100 mM solutions (Cat. No. 1 277 057); creatine kinase from rabbit muscle (Cat. No. 127 566); creatine phosphate, disodium salt (Cat. No. 127 574); and proteinase K (Cat. No. 1092766) are from Roche. Tris (Cat. No. 108382); KCl (Cat. No. 104936); KOH (Cat. No. 105021); MgCl2 (Cat. No. 105833); NaN3 (Cat. No. 822335); 25% NH3 solution (Cat. No. 105432); ethanol (Cat. No. 100983); and sodium salicylate (Cat. No. 106602) are from Merck. Spermidine (Cat. No. S 0266); bovine serum albumin, essentially fatty acid free (BSA) (Cat. No. A-7511); dithiothreitol (DTT) (Cat. No. D 5545); HEPES (Cat. No. H 7523); trypsin (Cat. No. T 1426); trypsin inhibitor, from soybean (Cat. No. T 9003); α-nicotinamide adenine dinucleotide disodium salt, reduced form (NADH) (Cat. No. N 6879); EDTA (Cat. No. E 9884); (NH4)2SO4 (Cat. No. A 2939); CaCl2, dihydrate (Cat. No. C 5080); magnesium acetate tetrahydrate (Cat. No. M 2545); valinomycin (Cat. No. V 0627); ATP, disodium salt (Cat. No. A 7699); glycerol (Cat. No. G 6279); potassium acetate (Cat. No. P 5708); KH2PO4 (Cat. No. P 5379); L-methionine (Cat. No. M 9625); urea (Cat. No. U 5128); phenylmethylsulfonyl fluoride (PMSF) (Cat. No. P 7626); tRNA, from bovine liver (Cat. No. R 4752); and Triton X-100 (Cat. No. T 9284) are from Sigma. Rabbit reticulocyte lysate (Cat. No. L 4960) and amino acid mixture, minus methionine (Cat. No. L 4960), are from Promega. NaCl (Cat. No. 9265.1) and trichloroacetic acid (TCA) (Cat. No. 8789.1) are from Roth. Sorbitol (Cat. No. 2039) is from Baker. L-[35S]methionine, >1000Ci/mmol (Cat. No. SJ 235), mWG(5Ç)ppp(5(Ç)G (G-cap) (Cat. No. 27 4635 02), and Kodak X-OMAT X-ray film (Cat. No. V1651496) are from Amersham Biosciences. Sorvall centrifuge RC M120 GX, Kendro. Sorvall Rotor S100 AT3-204, Kendro. Eppendorf centrifuge 5417 R, "microfuge" Eppendorf. Greiner PP-tubes 15ml (Cat. No. 188261), Greiner. X-ray cassettes (Cat. No Rö 13), GLW. Plasmid is pSP65mdh1 (Dubaquié et al., 1998). Highly purified mitochondria (25 mg/ml) are prepared after Glick and Pon (1995).
III. PROCEDURES
Solutions used directly as obtained from the supplier are only listed in Section II. Protocols for the preparation of solutions used throughout the procedure are only given once.
A. Transcription Using SP6 Polymerase
Solutions
- 1M Tris-HCl stock solution, pH 7.5: Dissolve 12.1 g Tris in 80ml H2O and adjust pH to 7.5 with 5M HCl. Add H2O to 100ml. Autoclave and store at room temperature.
- 1M HEPES-KOH, pH 7.4: Dissolve 23.8 g HEPES in 80ml H2O and adjust pH to 7.4 using 4M KOH. Add H2O to 100ml. Autoclave and store at room temperature.
- 1M spermidine: Dissolve 145 mg spermidine in 1 ml H2O. Store at -20°C.
- 100 mg/ml BSA: Dissolve 500 mg BSA in 5 ml H2O. Store at -20°C.
- 2.5M MgCl2: Dissolve 50.8 g MgCl2 in 100ml H2O. Autoclave and store at 4°C.
- 2.5 M KCl: Dissolve 18.6 g KCl in 100 ml H2O. Autoclave and store at 4°C.
- 100 mM DTT: Dissolve 15.4 mg DTT in 1 ml H2O. Store at -20°C. Make a fresh solution about every 4 weeks.
- 5× SP6 reaction buffer: 200mM Tris-HCl, pH 7.5, 30mM MgCl2, 10mM spermidine, and 0.5mg/ml BSA. To obtain 10ml of a 5× reaction buffer, mix 2 ml 1M Tris-HCl, pH 7.5, 120 µl 2.5M MgCl2, 100µl 1M spermidine and 50µl 100mg/ml BSA. If necessary, readjust the pH to 7.5. Store in 1-ml aliquots at -20°C.
- G-cap (m7G(5')ppp(5')G): Dissolve 25 A250 units in 242 µl H2O. Freeze 10-µl aliquots in liquid nitrogen. Store at -70°C.
- 5 mM NTP-GTP: To make a 500µl stock, add 25 µl 100 mM ATP, 25 µl 100 mM UTP, and 25 µl 100 mM CTP to 425 µl 20mM HEPES-KOH, pH 7.4. Store in 100-µl aliquots at -70°C.
- 5mM GTP: Mix 475µl 20mM HEPES-KOH, pH 7.4, with 25 µl 100 mM GTP solution.
- RNase inhibitor buffer: 20 mM HEPES-KOH, pH 7.4, 50mM KCl, 10mM DTT, and 50% glycerol. Make 10ml of the buffer by mixing 200µl 1M HEPES-KOH, pH 7.4, 200µl 2.5M KCl, 1 ml 100mM DTT, and 5 ml glycerol. Add H2O to 10ml and store at -20°C.
- 4 units/µl RNase inhibitor: Add 500µl RNase inhibitor buffer to 2000 units of RNase inhibitor. Store at -20°C for up to 6 months.
- 1µg/µl linearized plasmid DNA: Prepare the linearized plasmid (pSP65mdh1) according to standard molecular biology procedures.
Steps
- Mix the following solutions carefully, avoiding
the formation of air bubbles. Follow the indicated
order of addition because the DNA might precipitate
in 5x SP6 buffer. Precipitation of DNA can also occur
if the mixture is placed on ice. Incubate the mixture at
40°C for 15 min.
H2O 12µl 4 units/µl RNase inhibitor 1µl 1µg/µl linear plasmid (pSP65mdh1) 5µl 5 mM rNTPs minus GTP 5µl 5 mM G-cap 5µl 100 mM DTT 5µl 5× SP6 buffer 10µl SP6 polymerase 2µl
- Start transcription by adding 5 µl 5 mM GTP solution and incubate for 90 min at 40°C.
- Extract the mRNA with phenol/chloroform and then with chloroform/isoamylalcohol, precipitate with 100% ethanol, and wash with 70% ethanol. Resuspend the dried pellet in 125 µl H2O.
- The mRNA obtained by this procedure is used directly in the translation protocol, mRNA can be stored in 10-µl aliquots at -70°C. If frozen mRNA is used for translation, thaw rapidly and keep at room temperature before adding the mRNA to the translation reaction.
B. Translation Using Reticulocyte Lysate
Solutions
- 1M DTT: Dissolve 154mg DTT in 1 ml H2O. Store at -20°C. Make a fresh solution about every 4 weeks.
- 8mg/ml creatine kinase: Dissolve 8mg creatine kinase in 475 µl H2O. Add 20 µl 1M HEPES-KOH, pH 7.4, 5µl 1M DTT, and 500µl glycerol. Freeze in 10-µl aliquots in liquid nitrogen and store at -70°C.
- 5mg/ml tRNA: Dissolve 10mg tRNA from bovine liver in 2 ml H2O. Store in 100-µl aliquots at -20°C.
- 400mM HEPES-KOH, pH 7.4: Mix 6ml H2O with 4 ml 1M HEPES-KOH, pH 7.4. If necessary, readjust pH.
- 10 mM GTP: Mix 450 µl 20 mM HEPES-KOH, pH 7.4, with 50µl 100mM GTP. Store at -20°C.
- 100mM ATP: Dissolve 55.1mg ATP in 900µl H2O. Adjust to pH -7 using 4M NaOH and pH indicator paper. Adjust volume to 1 ml and store at -20°C.
- 600 mM creatine phosphate: Dissolve 153.06 mg creatine phosphate in 1 ml H2O. Store at -20°C.
- 4M potassium acetate: Dissolve 3.92g potassium acetate in 10 ml H2O. Do not adjust the pH. Store at -20°C.
- 50 mM magnesium acetate: Dissolve 10.7 mg magnesium acetate tetrahydrate in 1 ml H2O. Store at -20°C.
Steps
- Prepare the reticulocyte lysate mix and the tRNA
mix fresh. To obtain a 100-µl translation reaction, mix
the following solutions.
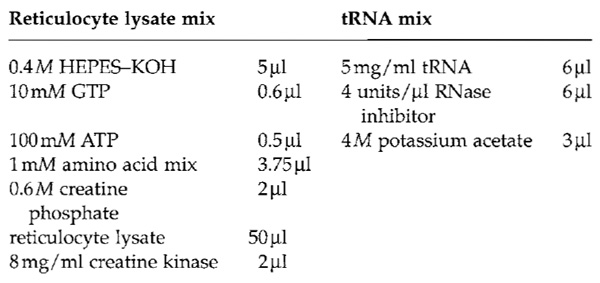
- Use the mRNA obtained in Section III,A. It is possible to use mRNA produced in a different transcription system, e.g., with T7 RNA polymerase. Mix 60 µl reticulocyte lysate mix, 10 µl tRNA mix, 2 µl 50 mM magnesium acetate, 18 µl mRNA, 10 µl [35S]methionine, and 2µl 1M DTT.
- Incubate this mixture for 60min at 30°C. Shield it from light to prevent heme-induced photooxidation of the precursor proteins. Remove ribosomes after the translation reaction by centrifugation for 15min at 150,000g (65,000rpm with S100 AT3-204 rotor in Sorvall centrifuge). Remove the supernatants, being careful not to disturb the ribosomal pellet.
C. Denaturation of Radiolabeled Precursor Protein
Solutions
- Saturated (NH4)2SO4 solution: Weigh 100 g (NH4)2SO4 and add H2O to a final volume of 100ml. Stir for 30 min at room temperature. The (NH4)2SO4 will not dissolve entirely. Remove the supernatant and keep at room temperature.
- 8M urea: Dissolve 4.85g urea in a final volume of 10ml 25 mM Tris-HCl, pH 7.5, containing 25 mM DTT.
Steps
- Proteins synthesized in reticulocyte lysate are either folded or bound to chaperone proteins present in the lysate (Wachter et al., 1994). In order to unfold the protein prior to import, it can be precipitated by high concentrations of ammonium sulfate and subsequently denatured in 8M urea.
- Add 200µl of the (NH4)2SO4 solution to the 100-µl translation reaction. Mix well and allow precipitation of the protein for 30 min on ice. Collect precipitate by centrifugation in an Eppendorf centrifuge at 20,000 g for 10 min.
- Discard the supernatant and dissolve the pellet in 100 µl of 8 M urea solution. Keep the denatured precursor at room temperature for 10-30 min. This precursor solution is used for the import reaction (Section III,D) and preparation of the precursor standard (Section III,G).
D. Import of Denatured Radiolabeled Precursor Proteins
Solutions
For additional solutions required, see Sections III,A and III,B.
- 2.4M sorbitol: Dissolve 43.7 g of sorbitol in a final volume of 100ml H2O. Autoclave and store at 4°C.
- 1M KH2PO4: Dissolve 1.36 g KH2PO4 in 10 ml H2O. Filter sterilize and keep at room temperature.
- 1M HEPES-KOH, pH 7.0: Dissolve 23.8 g HEPES in 80ml H2O and adjust pH to 7.0 with 4M KOH. Add H2O to a final volume of 100ml. Filter sterilize and store at room temperature.
- 250 mM EDTA, pH 7.0: Resuspend 7.3 g of EDTA in 70ml H2O. Adjust pH to 7.0 using 5 M NaOH. Add H2O to a final volume of 100ml. Filter sterilize and keep at room temperature.
- 2× import buffer: 1.2M sorbitol, 100mM HEPES-KOH, pH 7.0, 100 mM KCl, 20 mM MgCl2, 5 mM EDTA, pH 7.0, 4 mM KH2PO4, 2 mg/ml BSA, and 1.5mg/ml methionine. To make 100ml of 2× import buffer, mix 50ml 2.4M sorbitol, 400µl 1M KH2PO4 solution, 4ml 2.5M KCl, 10ml 1M HEPES-KOH, pH 7.0, 0.8ml 2.5M MgCl2, 2ml 250mM EDTA, pH 7.0, 150 mg methionine, and 200 mg BSA. Adjust pH to 7.0 and add H2O to 100ml. Store at -20°C.
- 1× import buffer minus BSA: Prepare 2× import buffer, but without BSA. To obtain 1× import buffer minus BSA, mix 2ml 2× import buffer with 2ml H2O.
- 500mM NADH: Dissolve 35.5 mg of NADH in a final volume of 100µl 20mM HEPES-KOH, pH 7.0. Store at -20°C.
- 1 mg/ml valinomycin: Dissolve 2mg valinomycin in 2ml ethanol. Store at -20°C.
- Purified yeast mitochondria: 25mg mitochondrial protein/ml. Store at -70°C in 0.6M sorbitol, 20mM HEPES-KOH, pH 7.4, and 10mg/ml BSA. Thaw rapidly at 25°C immediately before the experiment. Do not refreeze. A detailed protocol of the purification procedure is given in Glick and Pon, (1995).
Steps
- Import into the matrix of mitochondria requires a membrane potential across the inner mitochondrial membrane. Therefore, the most thorough control for the specificity of an import reaction is to determine its dependence on a membrane potential. Adding ATP and the respiratory substrate NADH generates this potential. (Note that mammalian mitochondria cannot oxidize added NADH.)
- Perform two import reactions, one in the absence
and one in the presence of valinomycin. Preincubate
the import reaction in a 15-ml Greiner tube at 25°C for
1-2 min.
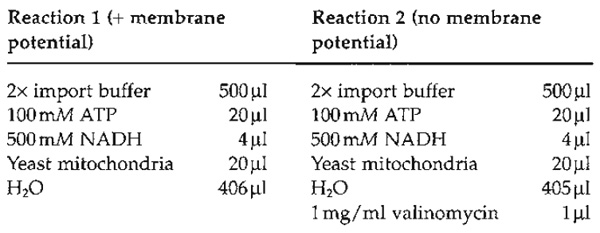
- Add 50µl of the denatured precursor protein solution (see Section III,C) containing denatured malate dehydrogenase to each reaction (reactions 1 and 2). Intact mitochondria should be handled gently. However, it is essential to mix the denatured precursor protein into the import reaction rapidly. Agitate the import reaction gently on a vortex mixer while adding the denatured precursor mixture dropwise. If mixing is performed only after addition, the precursor protein tends to aggregate and becomes import incompetent.
- Incubate at 25°C for 10 min. Agitate gently every other minute to facilitate gas exchange. Stop the import reaction by transferring the tubes onto ice. Add 1 µl of 1 mg/ml valinomycin to reaction 1.
- Remove 200µl each from reactions 1 and 2 and put the samples on ice. Spin down mitochondria in an Eppendorf centrifuge and remove the supernatant (be careful, the pellet will be very small). Resuspend the mitochondrial pellets of reactions 1 and 2 in each 200 µl of 1× import buffer. These samples represent the total of the two import reactions (Fig. 1, lanes 2 and 5). Add 22µl 50% TCA to each. Keep on ice and process further after all samples have been acid denatured (for the method of TCA precipitation, see Section III, G).
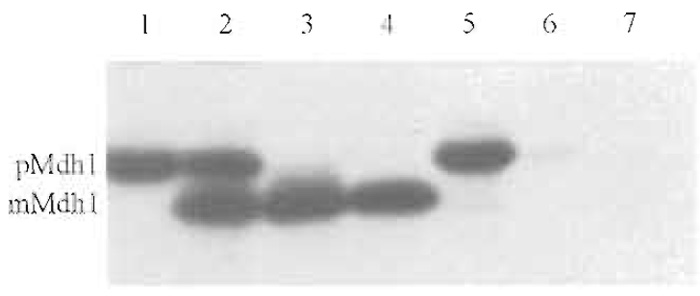 |
| FIGURE 1 Import of radiolabeled yeast malate dehydrogenase into isolated yeast mitochondria. Lane 1, 10% of the material added to each import reaction; lanes 2-4, import in the presence of ATP and a membrane potential across the inner mitochondrial membrane; and lanes 5-7, import in the absence of ATP and a membrane potential across the inner mitochondrial membrane. Lanes 2 and 5: total; material isolated together with the mitochondrial pellet. Lanes 3 and 6: import; material protease protected in intact mitochondria. Lanes 4 and 7: folded; material protease resistant even after solubilization of the mitochondria with Triton X-100. pMdh1, precursor form of Mdh1; mMdh1, mature form of Mdh1. For experimental details, see text. |
E. Protease Treatment of Intact Mitochondria
Solutions
- 10mg/ml trypsin: Dissolve 3 mg of trypsin in 300µl H2O. Make fresh.
- 20mg/ml trypsin inhibitor: Dissolve 6 mg of trypsin inhibitor in 300 µl H2O. Make fresh.
Steps
Perform the following steps in parallel with both import reactions.
- To digest precursor proteins that stick to the surface of the mitochondria, add 8µl 10mg/ml trypsin (final concentration 100µg/ml). Incubate for 30min on ice.
- Add 8µl 20mg/ml trypsin inhibitor (final concentration 200µg/ml) and incubate on ice for 5min.
- Transfer the sample into a new Eppendorf tube.
- Spin for 3 min in an Eppendorf microfuge at 10,000 g. Remove the supernatant carefully by aspiration.
- Carefully resuspend the mitochondrial pellet in 800µl 1× import buffer minus BSA. As it is extremely important to resuspend the pellet completely, it should be done as follows. First add 100 µl of l x import buffer minus BSA and resuspend mitochondria by pipetting up and down. Then add another 700µl of 1× import buffer minus BSA to yield 800 µl final volume.
- Remove 200µl of each sample and add 22µl 50% TCA. Keep on ice. These samples represent the material that has crossed the outer membrane completely (import, Fig. 1, lanes 3 and 6).
F. Inherent Protease Resistance of the Imported Protein
Solutions
- 1M Tris-HCl stock solution, pH 8.0: Dissolve 12.1 g Tris in 80ml H2O and adjust pH to 8.0 using 5M HCl. Add H2O to a final volume of 100ml. Autoclave and store at room temperature.
- 2 M CaCl2: Dissolve 14.7 g CaCl2 in 50 ml H2O. Autoclave and store at room temperature.
- 10% Triton X-100 (w/v): Dissolve 10g of Triton X-100 in a final volume of 100ml H2O. Store at room temperature in the dark.
- 10% NaN3: Dissolve 1 g NaN3 in a final volume of 10ml H2O. Store at room temperature.
- Proteinase K buffer: 50mM Tris-HCl, pH 8.0, 1 mM CaCl2, and 0.02 % NaN3. To make 10ml of proteinase K buffer, mix 500µl 1M Tris-HCl, pH 8.0, 5µl 2M CaCl2, and 20µl 10% NaN3. Add H2O to a final volume of 10ml. Store at room temperature.
- 10mg/ml proteinase K stock: Dissolve 5mg of proteinase K in 500 µl proteinase K buffer. Store at 4°C for up to 1 week without loss of activity.
- 200µg/ml proteinase K solution: Mix 10µl of 10mg/ml proteinase K stock with 390 µl H2O. Add 100 µl 10% Triton X-100. Make fresh.
- 200mM PMSF: Make a fresh solution of PMSF by dissolving 34.85 mg of PMSF in 1 ml of ethanol.
Steps
- Transfer 200 µl from the remainder of the import reaction into a fresh Eppendorf tube. Add 200 µl of the 200-µg/ml proteinase K solution and mix rapidly. Leave the tube on ice for 15 min.
- Add 2µl 200mM PMSF while agitating on a vortex mixer. Keep on ice for 5min. Add 44µl 50% TCA. Add 300µl of acetone to dissolve the Triton X- 100 that precipitates in the presence of TCA. These samples measure the fraction of the precursor protein that has completely crossed the outer membrane and has reached the folded state (folded, Fig. 1, lanes 4 and 7).
G. Final Processing of Samples and Preparation of a Precursor Standard Steps
- To inactivate proteases, incubate the TCAprecipitated samples (total, import, folded) at 65°C for 5 min. Place on ice for 5 min and subsequently collect the TCA precipitate by spinning for 10min at 20,000g.
- Remove supernatant by aspiration and dissolve the pellets in 30 µl 1× sample buffer. If the sample buffer turns yellow, overlay the sample with NH3 gas taken from above a 25% NH3 solution. Agitate to mix the gaseous NH3 gas into the sample buffer until the color turns blue again.
- Incubate the samples for 5 min at 95°C.
- To estimate the efficiency of the import reaction, the amount of precursor protein added to the import reaction has to be determined. The efficiency of import for most precursor proteins is between 5 and 30%. Here we use a 10% standard (Fig. 1, lane 1).
- To obtain a 10% standard, mix 4µl of purified yeast mitochondria (see Section III,D) with 30µl 1× sample buffer. Incubate at 95°C for 3 min.
- Add 1 µl of the precursor protein solution (Section III,C) and incubate for 5 min at 95°C.
H. SDS-Gel Electrophoresis and Processing of the Gel
Solutions
- 5% TCA: To make 5 liter, add 250g of TCA to 5 liter H2O.
- 1M Tris base: Dissolve 121 g of Tris in 1 liter H2O.
- 1M sodium salicylate: Dissolve 160 g of sodium salicylate in 1 liter H2O.
Steps
- Run samples on a 10% Tris-tricine gel (Schägger and von Jagow, 1987) stabilized by the addition of 0.26% linear polyacrylamide prior to polymerization.
- To reduce radioactive background, boil 5% TCA in a beaker under the hood. Add the gel to the boiling TCA and incubate for 5 min.
- Recover the gel and place it into a tray. Wash briefly with water. Neutralize by incubation in 1M Trisbase for 5 min on a shaker.
- Wash briefly with water. Add 1M sodium salicylate and incubate for 20min on a shaker.
- Dry gel on a Whatman filter paper and expose to a Kodak X-OMAT X-ray film for the desired time. Exposure time for the experiment shown in Fig. 1 was 12 h.
IV. COMMENTS
The method describes a standard experiment to test a precursor protein that has not been used in mitochondrial import before. Most importantly, as demonstrated here for malate dehydrogenase, the protocol will reveal if import is dependent on a membrane potential (compare Fig. 1 lanes 3 and 6). This is essential, as sometimes protease-resistant precursor proteins tend to stick to the outside of mitochondria, thereby "mimicking" import.
The efficiency of import can be deduced by a comparison of the amount of imported material with a precursor standard (compare Fig. 1 lanes 1 and 3). In addition, the experiment reveals if a precursor protein folds to a protease-resistant conformation after its import into the mitochondrial matrix. Under the conditions chosen here, complete protease resistance was obtained for malate dehydrogenase (Fig. 1, lanes 3 and 4).
V. Pitfalls
The quality of the DNA used for transcription is essential for efficiency. Use a clean, RNA- and RNasefree plasmid preparation (e.g., purified with a Qiagen plasmid kit, Qiagen). Linearize plasmid by cutting with a restriction enzyme behind the coding region of the gene of interest. Extract with phenol/chloroform and then with chloroform/isoamylalcohol, precipitate with 100% ethanol, and wash with 70% ethanol. Resuspend the dried pellet in H2O at a concentration of 1 µg/µl and store at 4°C. Never freeze DNA templates used for transcription.
To avoid RNase contamination, solutions used for transcription and translation have to be prepared with special caution. Always wear gloves even when loading pipette tips into boxes. If initiation at downstream AUG codons is a problem, try diluting the reticulocyte lysate up to fourfold.
It is important to establish that import is linear with time. To establish those conditions it is necessary to perform time course experiments of the import reaction and to try import at different temperatures.
Methods for determining the intramitochondrial localization of an imported precursor protein (Glick, 1991), investigating the energy requirements of mitochondrial import (Glick, 1995), and detecting interaction between imported precursor proteins and matrix chaperones (Rospert and Hallberg, 1995) have been published elsewhere.
References
Dubaquié, Y., Looser, R., Fünfschilling, U., Jenö, E, and Rospert, S. (1998). Identification of in vivo substrates of the yeast mieochondrial chaperonins reveals overlapping but non-identical requirement for hsp60 and hspl0. EMBO J. 17, 5868-5876.
Glick, B. S. (1991). Protein import into isolated yeast mitochondria. Methods Cell Biol. 34, 389-399.
Glick, B. S. (1995). Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae. Methods Enzymol. 260, 224-231.
Glick, B. S., and Pon, L. A. (1995). Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260, 213-223.
Jensen, R. E., and Yaffe, M. P. (1988). Import of proteins into yeast mitochondria: The nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MASl-encoded subunit. EMBO J. 7, 3863-3871.
Melton, D. A., Krieg, E A., Rebagliati, M. R., Maniatis, T., Zinn, K., and Green, M. R. (1984). Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12, 7035-7056.
Neupert, W. (1997). Protein import into mitochondria. Annu. Rev. Biochem. 66, 863-917.
Pfanner, N., and Geissler, A. (2001). Versatility of the mitochondrial protein import machinery. Nature Rev. Mol. Cell. Biol. 2, 339-349.
Rospert, S., and Hallberg, R. (1995). Interaction of HSP 60 with proteins imported into the mitochondrial matrix. Methods Enzymol. 260, 287-292.
Sch/igger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel elektrophoresis for the separation of proteins in the range from 1 to 100kDa. Anal. Biochemi. 166, 368-379.
Wachter, C., Schatz, G., and Glick, B. S. (1994). Protein import into mitochondria: The requirement for external ATP is precursorspecific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol. Biol. Cell. 5, 465-474.




