Addition of caps (m7G) and tails (polyA) for mRNA
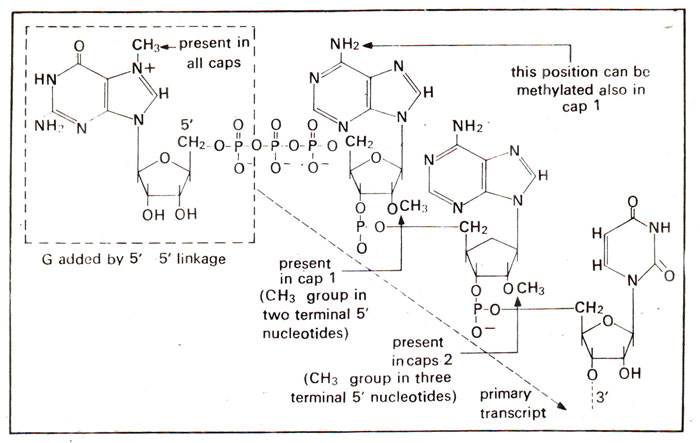
Fig. 33.1. Capping of 5' end of mRNA by cap O, cap 1 and cap 2 due to methylation at several positions.
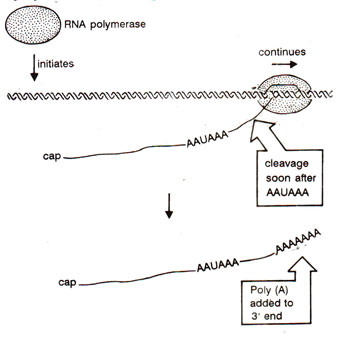
Fig. 33.2. Cleavage at AAUAAA and polyadenylation at the cleaved 3’ end.
The initial RNA transcript (hnRNA), derived from genes coding for proteins, gets modified so that its 5' end gains a methylated guanine (caps) and its 3' end is polyadenylated (tails). Capping at 5' end occurs rapidly after the start of transcription and much before completion of transcription. Transcription starts with a nucleoside triphosphate (A or G), and a 5' triphosphate group is retained at this first position. The initial sequence at 5' end of hnRNA is therefore 5' ppp A(or G)pNpNp...3'. To the 5' end is added a terminal G with the help of an enzyme guanyl transferase as follows :
5' Gppp + 5' ppp A (or G) pNpNp → 5' Gppp 5'ApNpNp + pp + p
The new G residue is in the reverse orientation with respect to all other nucleotides and undergoes methylation (using a methyl transferase at its 7th position.

Fig. 33.1. Capping of 5' end of mRNA by cap O, cap 1 and cap 2 due to methylation at several positions.

Fig. 33.2. Cleavage at AAUAAA and polyadenylation at the cleaved 3’ end.
It could be shown that removal of cap leads to loss of translation activity due to loss of the formation of mRNA-ribosome complex. It suggests that the 'cap' helps in recognition of ribosome. Only in some eukaryotic mRNAs (as those for histone proteins), caps may be absent and may not be required for translation.
The 3' end of mRNA is generated in two steps (Fig. 33.2) (i) Nuclease activity cuts the transcript at an appropriate location, (ii) Poly (A) is added to the newly generated end by an enzyme, poly (A) polymerase, utilizing ATP as a substrate.
Estimations have shown that ordinarily ~30% of hnRNA and ~70% of mRNA are polyadenylated. In a region 11-30 nucleotides upstream of the site of poly (A) addition, there is a sequence AAUAAA (in all higher eukaryotes except yeast), which perhaps gives a signal for cleavage (step (i) above). In most histone proteins mRNAs, no polyadenylation at 3' end is found, so that 3' ends are processed without addition of poly (A). The U7 snRNA (56 bases long and not related with other snRNAs involved in spliceosome; see later) is involved in this processing, through extensive complementary base pairing with histone mRNAs (or hnRNAs). Other proteins may also be involved in processing of 3' end of hnRNA, whether or not it is polyadenylated.




