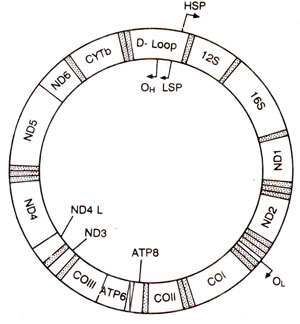
Fig. 32.28. A map of vertebrate mtDNA, showing location of D loop, origins for DNA replication (Oh and Ol), promoters for transcription (HSP and LSP), genes for 22 tRNAs (shaded areas) and other protein coding genes (CO = cytochrome oxidase; ATP6, ATP8 = ATPase subunits; cyt b = cytochrome b; ND = NADH dehydrogenase); arrow indicate direction of replication or transcription.
The transcription of each strand of vertebrate mtDNA starts at a site within the
D loop ('displacement loop', where a short piece of nascent DNA is found at the heavy or H strand). The transcription continues uninterrupted, so that in the transcript, tRNA sequences lie among other RNA sequences. During RNA processing, tRNA species and different mature mRNAs are produced. The most abundant RNA species in mitochondria are, however, two rRNA species, which are transcribed separately and at a faster rate. In human and mouse mtDNA, the two promoters for transcription occur within the D loop, separated from one another by 150bp (Fig. 32.28). These are called
H-strand promoter (HSP) and
L-strand promoter (LSP). Each promoter (5Obp) has two functional domains : (i) a short sequence at the transcription start site (which is absolutely necessary for transcription initiation), and (ii) an upstream sequence- (-12 to -39), which increases efficiency of transcription initiation. A transcription factor, called
mtTF-1 binds to the sequence upstream to 'start site' and has greater affinity for LSP than for HSP. The species-specific transcription is, however, determined either by mtRNA polymerase (mtRNAP), or by some frans-acting factor(s) or both.
The mtTFl activates transcription by inducing physical changes (unwinding and bending of DNA) at the promoter, in concert with other proteins essential for transcription. The mtRNAP later binds at the start site and initiates transcription. The protein, mtTF-1 has been isolated and purified from human and mouse. A similar protein has also been isolated from yeast mitochondria. Nuclear genes encoding yeast mtRNAP and human mtTF-1 have recently been cloned and sequenced. The mtRNAP is a single polypeptide like that in the RNA poiymerases of T3 and T7 phages and differs from bacterial RNA polymerase, having four polypeptides (α
2ββ
’).

Fig. 32.28. A map of vertebrate mtDNA, showing location of D loop, origins for DNA replication (Oh and Ol), promoters for transcription (HSP and LSP), genes for 22 tRNAs (shaded areas) and other protein coding genes (CO = cytochrome oxidase; ATP6, ATP8 = ATPase subunits; cyt b = cytochrome b; ND = NADH dehydrogenase); arrow indicate direction of replication or transcription.
Although, nothing is known about the events associated with elongation of mitochondrial transcripts, but some progress in our understanding of the mode of termination of transcription has been made for human mtDNA. A 13bp sequence (5'-TGGCAGAGCCCGG-3') has been found to be adequate to direct termination. A termination protein is perhaps involved, which binds at the termination site and acts as a barrier to the transcription process.






