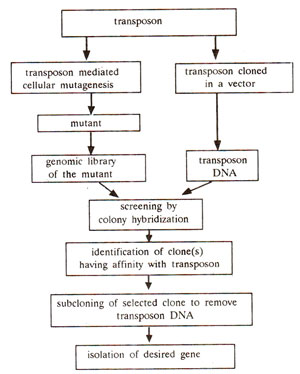
Fig. 41.4. Steps involved in the use of transposon mutagenesis (or tagging) for identification and isolation of a gene.
In some cases, we are interested in isolation of a gene whose phenotypic effect is known, but the gene product has not been identified or can not be isolated. Such genes include those for some morphological traits, dormancy, photoperiodicity, disease resistance, etc. This area of research, in which genetics is studied by isolating the gene first without knowing the gene product is often described as
reverse genetics (see
Genetics : An Overview and
Genetic Engineering and Biotechnology 1. Recombinant DNA and PCR (Cloning and Amplification of DNA)). The methods used for the isolation of 'these genes are different from those used for genes coding for known proteins. Some of these methods are described in this section.
Use of transposable elements (transposon or gene tagging). The transposable elements (TEs), in some cases, have been effectively utilized for isolation of genes, when the gene product is unknown. In this case a transposon works as a mutagen and therefore as a gene tag. Following steps which are illustrated in Figure 41.4, are involved in this procedure : (i) A gene with a scorable phenotypic effect is cloned, (ii) TE is transposed to this gene to get an unstable allele. (iii) This unstable allele is cloned and TE is isolated from this unstable allele (this is to select a TE which can produce unstable allele). (iv) This TE is transposed to a gene of interest with known phenotypic effect, to produce unstable allele. (v) The DNA is extracted from this mutant, (vi) TE sequence is used as a probe to isolate and clone the mutant gene (carrying inserted TE), so that we can then isolate the gene of interest.

Fig. 41.4. Steps involved in the use of transposon mutagenesis (or tagging) for identification and isolation of a gene.
In maize, TE like Ac/Ds, En/Spm and Mu1 have been isolated using the genes
Wx, C2 and
Adh1. These TEs can be used for gene tagging experiments leading to isolation of genes. In maize, several genes like
Bz1, P, A1, C1 and
C2 have been isolated successfully using gene tagging method.
For gene tagging, often transposable elements endogenous to species like maize and snapdragon have been used. However, rarely transposdhs available from one plant can be moved into the genome of another plant, whose gene is to be isolated. For instance Ac element of maize has been transferred to tobacco, where it can integrate into any locus permitting gene tagging and gene isolation.
Mutant complementation. In this technique, clones from the wild type strain are selected which should be able to complement the mutants transforming them into wild type. Using these clones, the protoplasts derived from the mutant plant may be transformed and the transgenic plants are produced. Once this is done, the gene of interest can be isolated from DNA extracted from the transformed plants using wild type complementary clone as a probe.
Use of RFLP maps or chromosome walking for gene isolation. In recent years RFLP (restriction fragment length polymorphism) maps have been prepared in maize, tomato, rice, etc. Linkage of RFLP loci with genes of interest are also being worked out as an aid to plant breeding (see
Genetic Engineering and Biotechnology 2. Restriction Maps and Molecular Genetic Maps). If RFLPs are known which are very close to the target gene (as may be done through RFLP maps or chromosome walking), then by locating the RFLPs on either side of the target gene, a long intervening DNA segment may be isolated. This fragment may be used for subcloning leading to the isolation of the desired gene. If RFLPs are examined in two isogenic lines differing for a single specific gene, the difference in RFLP maps of these two lines will help in locating the position of this gene on molecular map which can then be utilized for isolation of the gene.
Use of chromosome jumping for gene isolation. In
Genetic Engineering and Biotechnology 1. Recombinant DNA and PCR (Cloning and Amplification of DNA), we described the technique of chromosome jumping for walking long chromosome distances, thus bringing the available known sequence (related to the available probe) closer to the gene of interest. Once this has been achieved, the technique described in the previous section may be conveniently used for isolating this gene for further study.






