Minisatellites (VNTRs) and Microsatellites (SSRs)
It has also been recently emphasized that polyallelic markers will be very useful for mapping both the simple Mendelian traits as well as polygenic traits in segregating populations. The most widespread of these polyallelic markers are minisatellites or the variable number of tandem repeat (VNTR) loci, which are uncovered by locus specific probes and exhibit highly polyallelic fragment length variation.
An interesting and common example of VNTRs involves the number of tandem repeat loci associated with rRNA genes in rDNA concentrated at nucleolar organizing regions (NORs) of specific chromosomes of an organism. Like rRNA genes, most VNTR loci are concentrated in proterminal regions of chromosomes, and thus may not be able to provide the desired density of markers. This problem has largely been overcome in microsatellite or SSR loci.
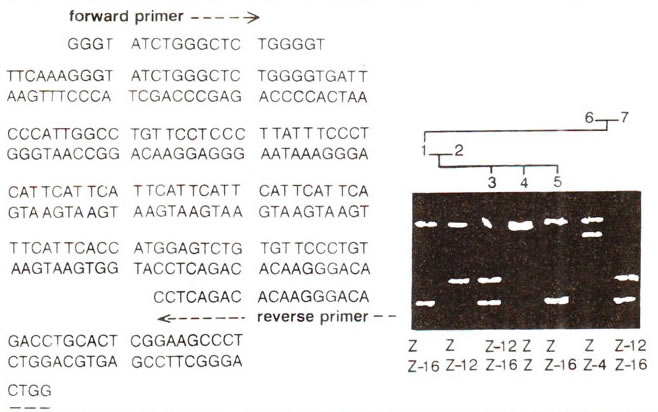
Microsatellites or simple sequence repeats (SSRs) represent variation in the repeat number of sequences, 1-6 bp long (VNTRs can be as long as 1 kbp), such as (AC)n or (AAC)n, etc. Being smaller in size, SSRs are more common and amenable to PCR analysis (Fig. 40.10), Recently, more then 300 loci representing microsatellites, were mapped each in human and mouse genomes. Similar effort are also being made in several crop plants.
Microsatellites or simple sequence repeats (SSRs) represent variation in the repeat number of sequences, 1-6 bp long (VNTRs can be as long as 1 kbp), such as (AC)n or (AAC)n, etc. Being smaller in size, SSRs are more common and amenable to PCR analysis (Fig. 40.10), Recently, more then 300 loci representing microsatellites, were mapped each in human and mouse genomes. Similar effort are also being made in several crop plants.