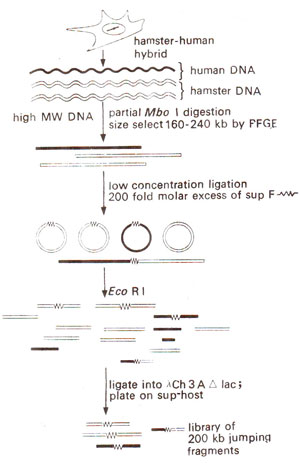
Reverse Genetics and Chromosome Jumping (or Hopping) Libraries
In most cases, we identify the genes by their protein products, which help in the isolation and cloning of a specific gene. However, for many-human disorders (e.g. huntington disease, cystic fibrosis, duchenne muscular dystrophy, etc.), the gene product is not known. Such genes are cloned by a process called 'reverse genetics', in which the gene is identified primarily by its map position. (For a new definition of reverse genetics by P. Berg, see page8). Once the gene is cloned, it can be utilized for getting information about the encoded protein, the gene mutation and the metabolic delect that may be associated with the disease. Since the information about the genetics is coming in the reverse order, this is popularly described as reverse genetics.
The reverse genetics approach involves the following two steps : (i) locate the gene to a particular chromosome through RFLP linkage analysis, (ii) map the position of the gene with respect to molecular marker, both through recombinational analysis (in terms of recombination value in cM = centiMorgan units) and by physical mapping (in terms of kilobase pairs) with pulsed-field gel electrophoresis (PFGE) i.e. through the preparation of restriction map. Even if the distance is IcM, which is the limit of resolution, this will be equivalent to >l,000kb. Therefore if the segment carrying the gene is to be cloned using the linked molecular marker, such a segment will be too large to be cloned. In order to bring the molecular marker close to the gene of interest, 'chromosome jumping' approach has been utilized.
The technique of 'chromosome jumping' is based on the following steps (Fig. 40.12) : (i) depending upon the distance between the gene and the marker, decide about the distance of 'jumps' or 'hopsize' (e.g. 100kb or 200kb); (ii) genomic DNA molecules in the range of selected size (say 80kb-130kb in case of 100kb 'hopsize' or 160-240kb for 'hopsize' of 200kb) are selected through pulsed-field gel electrophoresis; (iii) for circularization of DNA segments, ligation between two ends of each long linear DNA molecule was allowed using T4 ligase in the presence of sup F, (sup F is a suppressor tRNA gene that will allow selection of clones representing junction fragments by cloning them in an amber mutated phage sup F-); (iv) DNA circles obtained in (iii) above are digested with EcoRI; (v) the vector λCh3A Δ lac, an amber-mutated phage vector (sup F-) is also cut with EcoRIand used for cloning small DNA fragments representing the junctions of the circularized genomic DNA molecules and carrying sup F+; (vi) the cloned DNA fragments obtained in (v) above represent the jumping library, which can be plated on a bacterial host and screened through the technique of plaque hybridization described earlier.
The above technique of chromosome jumping will help narrowing the gap between the gene and available molecular markers, so that after several cycles of chromosome jumping followed by cloning the regions that are closer to the gene, it will be possible to approach very close to the desired gene and clone it. It is thus obvious that in the field of 'reverse genetics' using technique of chromosome jumping, it will be possible to isolate, clone and characterize genes, whose gene products are unknown. The gene for Cystic fibrosis was isolated using this technique. It could also be shown that 'Cystic fibrosis' disease is caused by a single base substitution in this gene. This technique of 'reverse genetics' will become a very active area of research particularly in human genetics in the coming years. Paul Berg (Noble Laureate), in 1991 has redefined 'reverse genetics' (see Genetics : An Overview.