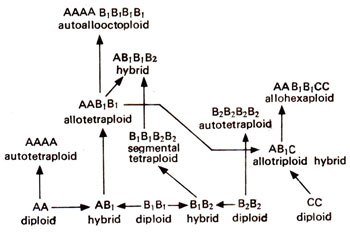
Polyploidy
There are mainly three different kinds of polyploids, namely (i) autopolyploids, (ii) allopolyploids, (iii)segmentalallopolyploids. Let us imagine that A1 B1 B2 and C are four different haploid sets of chromosomes and that genomes B1 and B2 are related. Different kinds of polyploids Using these genomes are derived in Figure 20.8.
Autopolyploids. Autopolyploids are those polyploids, which have the same basic set of chromosomes multiplied. For instance, if a diploid species has two similar sets of chromosomes or genomes (AA), an autotriploid will have three similar genomes (AAA) and an autotetraploid will have four such genomes (AAAA) as shown in Figure 20.8.
(a) Origin and production of autopolyploids.
The autopolyploids may occur in nature or may be artificially produced. When they are found in nature, their autopolyploid nature is inferred mainly by their meiotic behaviour. One of the very common examples of natural autopolyploidy relevant to Northern India, pertains to common 'doob' grass (Cynodon dactylon). In U.P. and Bihar, common 'doob' grass was found to be an autotriploid as inferred from its meiotic behaviour (Gupta and Srivastava, 1970).It is perhaps successful due to efficient vegetative reproduction, because, as will be seen, autoeupolyploids are normally triploids and set no seeds. Autotriploids are also known in watermelons, sugarbeet, tomato, grapes and banana, although in several of these cases the polyploids have been artificially produced. Similarly autotetraploids are known in rye (Secale cereale), corn (Zea mays), red clover (Trifolium pratense), berseem (Trifolium alexandrium), marigolds (Tagetes), snapdragons (Antirrhinum), Phlox, grapes, apples, etc. In Oenothera lamarckiana, an American plant, a giant mutant described by Hugo de Vries was later discovered to be an autotetraploid.
(b) Induced autopolyploids.
In many cases listed above, autopolyploidy is artificially induced. Since polyploids are normally larger and more vigorous, their role in crop improvement has been realized and techniques developed for artificial induction of polyploidy. Polyploidy is mainly induced by treatment with aqueous solution of a drug called colchicine. This drug has the property of arresting and breaking the spindle so that a cell division without cell wall formation may be affected leading to doubling of chromosome number. The concentration of aqueous solution of colchicine may vary from 0.01 % to 0.50% and the treatment may be given in one of the following manners, (i) Seed treatment may be mainly given by soaking seeds for different durations in. aqueous solution of colchicine. (ii) Injections of colchicine solution may also be given at seeding stage so as to inject solution into cortex tissue with the help of a hypodermic needle, (iii) Axiliary bud treatment is also effective. Since bud is meristematically active, placing cotton soaked in colchicine on the bud and continuous dropping of solution on the bud leads to induction of polyploidy in the branch arising from the treated bud. (iv) Shoot apex treatment is brought about just like bud treatment and is fairly effective, but the shoot apex should come in direct contact of the solution. In order to facilitate this, young leaves covering the shoot apex may be removed.
Colchicine is an alkaloid extracted from seed and corm of Colchicum autumnale. The action of colchicine and its use in inducing polyploidy was first studied in 1930's. The successful doubling of chromosome number was described for the first time by Levan (1938) and by Eigsti (1938-40). It was surprising however, that colchicine does not affect Colchicum, the plant from which it is extracted. This was attributed to the presence of an 'anticolchicine'. G.D.H. Bell (1959), who was once the Director of Plant Breeding Institute at Cambridge (England) had suggested that the success of colchicine treatment will depend on three prerequisites, namely (i) direct contact of growing tissue with solution, (ii) effective concentration of colchicine and (iii) the optimal stage of development. These three conditions are met with in the methods of treatment described above.
(c) Effects of chromosome doubling.
One of the important effects of polyploidy is to produce 'gigantism'. The autopolyploids may be normally larger in size. Sometimes plants may be smaller than diploids, but leaves, flowers and the cells themselves may be bigger in size. Some of the important effects of polyploidy are a's follows : (i) With increase in cell size, water content increases leading to decrease in osmotic pressure. This results into loss of resistance against frost, etc. (ii) Growth rate decreases due to slower rate of cell division; this leads to a decrease in auxin supply and a decrease in respiration, (iii) In autopolyploids, time of blooming is delayed and prolonged due to slow growth rate, (iv) At higher ploidy level (autooctoploids or higher), the adverse effects are highly pronounced and lead to death of plants.
(d) Cytology of autopolyploids.
In an autopolyploid, there will be more than two sets of homologous chromosomes. This leads to formation of multivalents instead of bivalents as found in diploids. An important difference exists even between autotriploids and autotetraploids, because while in the latter normal disjunction is possible giving rise to diploid gametes, in triploids it is not possible. In an autotriploid, there are three sets of homologous chromosomes. If these three sets are normally paired, trivalents can not disjoin normally and will either disjoin 2 : 1 chromosomes to two poles or will disjoin 1 : 1 leaving one chromosome as a laggard. The number of chromosomes in gametes of triploid organism, therefore, will vary from n to 2n. Most of these gametes are unbalanced leading to high degree of sterility.
In autotetraploids, since there are four sets of chromosomes, quadrivalents are formed, which disjoin in a normal 2 : 2 manner giving diploid gametes. Rarely however, a quadrivalent may disjoin 3 : 1 or may leave a chromosome as a laggard at anaphase I. Therefore autotetraploids also have a certain degree of sterility, although it will not be as high as in autotriploids.
(e) Genetics of autopolyploids (trisomic and tetrasomic inheritance).
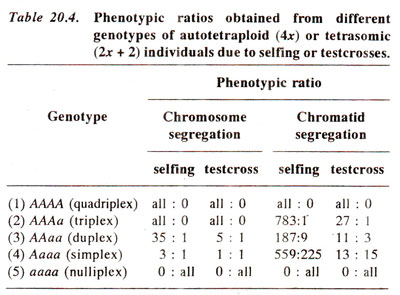
The segregation pattern in polyploids is quite different than what we find in diploids. In a polyploid there are more than one kind of heterozygotes, because there are more than two homologous chromosomes. The different possible genotypes and expected ratios in autotetraploids are given in Table 20.4. Similar ratios for triplqids are given in Table 20.1. The ratios are based on the assumption that either the gene is close to centromere leading to what we call chromosome segregation or the gene is away from centromere so that crossing over between gene and centromere will take place and ratios will be modified. In the latter case it will be called chromatid segregation, because segregation is taking place not at chromosome level, but at chromatid level due to crossing over.
It can be seen from Table 20.4 that main effect of autopolyploidy on segregation, when compared with that of diploids is the reduction in frequency of genotype homozygous at a particular locus. The result is that recessive phenotype is much less frequent as shown by a ratio 35 : 1 as against a disomic ratio of 3 : 1 and so on (Table 20.4). Tetrasomic inheritance and trisomic inheritance are useful criteria to differentiate between autopolyploids and allopolyploids and for locating genes on specific chromosomes.
(f) Uses of induced polyploidy
Since fertility level and seed set are low in induced polyploids, their utility in improvement of crop plants will depend on two major considerations, namely : (i) what is the commercial product, because if seed is not the commercial product low seed set will not be a handicap, and (ii) whether or not fertility level is important. Keeping these requirements in mind, seedless fruits can be produced by using triploids as in case of seedless watermelons produced by H. Kihara (who died in 1986) in Japan. These triploids were obtained from seeds raised by a cross tetraploid x diploid, the tetraploids being raised from diploids by colchicine treatment. This method was also used for obtaining triploids in sugarbeet, tomato and grapes.
Autotetraploids could also be used commercially. For instance, among grain crops autotetraploid rye is grown in Germany and Sweden. Tetraploids were also raised in barley and corn, although commercially they could not prove to be very useful. Among fruits, tetraploid apples and grapes have been found to be useful. Several ornamentals including marigolds, snapdragons, lily and Phlox are induced autotetraploids. Among forage crops, tetraploid berseem is a very popular crop in Northern India. In all cases of commercial uses of autotetraploids, fertility can be improved by selection, but sometimes unconsciously selection leads to reversion to diploidy and original tetraploid crop becomes diploid again. Recently D.R. Dewey (1980) in his article 'Some applications and misapplications of polyploidy in plant breeding' published in a book 'Polyploidy : Biological Relevance', emphasized that none of the autopolyploids mentioned above (triploid sugarbeet, tetraploids red clover and alsike clover, triploid watermelons and several ornamentals) lived upto the expectation. Although, if sustained efforts are made, some autopolyploids may prove to have breeding value, but plant breeders may question the time, money and labour spent towards this goal.
 Allopolyploids.
Allopolyploids.
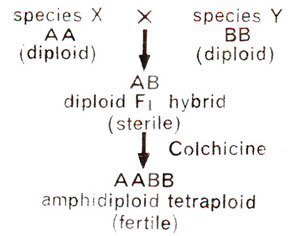
Polyploidy may also result from doubling of chromosome number in a F1 hybrid which is derived from two distinctly different species. This will bring two different sets of chromosomes in F1 hybrid. The number of chromosomes in each of these two sets may differ. Let A represent a set of chromosomes (genome) in species X, and let B represent another genome in a species Y. The F1 will then have one A genome and another B genome. The doubling of chromosomes in this F1 hybrid (AB) will give rise to a tetraploid with two A and two B genomes (Fig. 20.9). Such a polyploid is called an allopolyploid or amphidiploid.
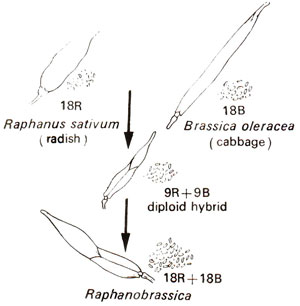
Raphanobrassica is a classical example of allopolyploidy. In 1927, G.D. Karpechenko, a Russian geneticist, reported a cross between Raphanus sativus (2n = 18) and Brassica oleracea (2n = 8) to produce F1hybrid which was completely sterile. This sterility was due to lack of chromosome pairing, since there is no homology between genomes from Raphanus sativus and Brassica oleracea. Among these sterile F1 hybrids, Karpechenko found certain fertile plants. On cytological examination these fertile plants were found to have 2n = 36 chromosomes, which showed normal pairing into 18 bivalents (Fig. 20.10).
(a) Evolution of wheat.
Common cultivated wheat is another important example of allopolyploidy, although its allopolyploid nature has now been questioned. There are three different chromosome numbers in the genus Triticum, namely 2n = 14, 2n = 28 and 2n - 42. The common wheat is hexaploid with 2n = 42, and is derived from three diploid species : (i) AA = Triticum aegilopoides (2n = 14), (ii) BB = Aegilops speltoides ? (2n = 14) (in the past evidence was made available, showing that Ae. speltoides may not be the donor of B genome; it is also believed that the donor of B genome may never be discovered) and (iii) DD = Aegilops squarrosq (2n = 14). The hexaploid wheat, therefore, is designated as AABBDD, the tetraploid (2n = 28) as AABB and diploid (2n = 14) as AA. There is, however, evidence available now which suggests that A, B and D genomes from three diploid species mentioned above are not much different from one another, so that it is believed that the three diploid progenitors of common hexaploid wheat were derived from a common ancestor. For this reason, the common hexaploid wheat is now considered an autopolyploid rather than an allopolyploid. At best, it may be a segmental allopolyploid.
(b) Synthesized allopolyploids.
Certain allopolyploids were artificially produced in order to find out the origin of naturally occurring allopolyploids. Common hexaploid wheat and tetraploid cotton furnish two such examples. These two and few other examples will be briefly discussed.
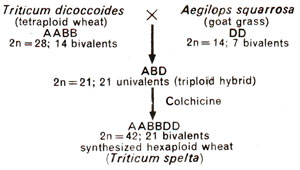
 (1) Triticum spelta, a hexaploid was artificially synthesized in 1946 by E.S. McFadden and E.R. Sears, and also by H. Kihara. They crossed an emmer wheat (tetraploid; 2n = 28) with Aegitops squarrosa (diploid; 2n = 14), and doubled the chromosome number in F1 hybrid (Fig. 20.11). The hexaploid or amphiploid synthesized in this manner was found to be similar to the primitive wheat T. spelta. When this synthesized hexaploid was crossed with naturally occurring T. spelta, F1 hybrid was completely fertile and showed normal pairing of chromosomes into bivalents. Therefore, it is suggested that hexaploid wheat must have originated in the past due to natural hybridization between tetraploid wheat and goat grass (Aegilops squarrosa)followed by subsequent chromosome doubling.
(1) Triticum spelta, a hexaploid was artificially synthesized in 1946 by E.S. McFadden and E.R. Sears, and also by H. Kihara. They crossed an emmer wheat (tetraploid; 2n = 28) with Aegitops squarrosa (diploid; 2n = 14), and doubled the chromosome number in F1 hybrid (Fig. 20.11). The hexaploid or amphiploid synthesized in this manner was found to be similar to the primitive wheat T. spelta. When this synthesized hexaploid was crossed with naturally occurring T. spelta, F1 hybrid was completely fertile and showed normal pairing of chromosomes into bivalents. Therefore, it is suggested that hexaploid wheat must have originated in the past due to natural hybridization between tetraploid wheat and goat grass (Aegilops squarrosa)followed by subsequent chromosome doubling.
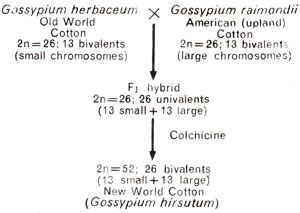
(2) Gossypium hirsutum, popularly known as upland cotton, is another interesting example of amphidiploidy. Old World cotton has 13 pairs of large chromosomes, while American or upland cotton has 13 pairs of smaller chromosomes. The New World cotton, the cultivated long-staple type has 26 paris of chromosomes, 13 large and 13 small. J.O. Beasley crossed the American and Old World cottons and doubled the chromosome number in the F1 hybrids (Fig. 20.12). The amphidiploid thus produced resembled the cultivated New World cotton and when crossed with it gave fertile F1 hybrids. The evidence presented in Figure 20.12, thus showed that New World cotton, the cultivated Gossypium hirsutum (tetraploid) originated from two diploid species, namely G. herbaceum (2n = 26) and G. raimondii (2n = 26).
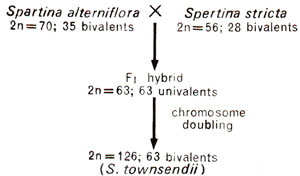
(3) Spartina townsendii, commonly called as Townsend's grass and found in Europe, has a chromosome number, 2n = 126. This species' in its morphology is intermediate to Spartina alterniflora (2n = 70), the American marsh grass and S. stricta (2n = 56), the European marsh grass. A study of morphology and cytology of these three species and the fact that S. townsendii was fertile, suggested that S. townsendii originated by natural hybridization of S. alterniflora and S. stricta, followed by doubling of chromosome number in the F1 hybrid so derived. This is thus an amphiploid (Fig. 20.13).
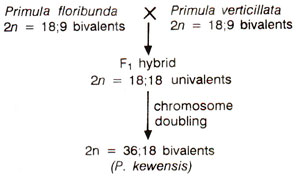
 (4) Primula kewensis is an allotetraploid (amphidiploid) with 2n = 4x = 36 and was derived from a Ax branch of a 2x hybrid between P. floribunda (n = x = 9) and P. verticillata (n = x = 9). Crosses can be easily made between P. floribunda and P. verticillata and the F1 hybrid with 2n = 18 is sterile showing almost complete lack of pairing. This difficulty is overcome by doubling of chromosome number (Fig. 20.14).
(4) Primula kewensis is an allotetraploid (amphidiploid) with 2n = 4x = 36 and was derived from a Ax branch of a 2x hybrid between P. floribunda (n = x = 9) and P. verticillata (n = x = 9). Crosses can be easily made between P. floribunda and P. verticillata and the F1 hybrid with 2n = 18 is sterile showing almost complete lack of pairing. This difficulty is overcome by doubling of chromosome number (Fig. 20.14).
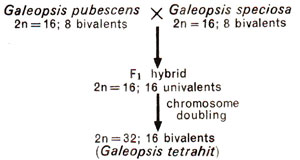
(5) Galeopsis tetrahit (2n = 32) could be synthesized (Fig. 20.15) from crosses involving diploid species G. pubescens (2n = 16) and G. speciosa (2n = 16). However, A. Muntzing, who performed these experiments suggested that G. tetrahit is not a strict amphidiploid, but has a tendency towards autopolyploidy.
The above examples indicate that a number of natural polyploids have originated by a process which can be duplicated in laboratory. Since the products obtained in laboratory resemble those found in nature, the method used in laboratory indicated evolutionary path which must have been followed in nature in remote past.
(c) Triticale (X Triticosecale), a new man made cereal.
In recent years, considerable emphasis has been laid on the possibility of utilizing a new man made cereal known as triticale, on a commercial scale. It is already grown in an estimated area of more than one million hectares and research at several places all over the world is in progress to improve this man made crop. Work done in this crop has been reviewed in detail (Muntzing, 1979, Gupta and Priyadarshan, 1982; Gupta and Reddy, 1991). Several hundred cultivars of triticale have been released during the last 10 years and more cultivars of this crop are being released every year. The area under its cultivation is also fast increasing (for more details consult, Gupta, 1984, 1985, 1987; Gupta and Reddy, 1991).
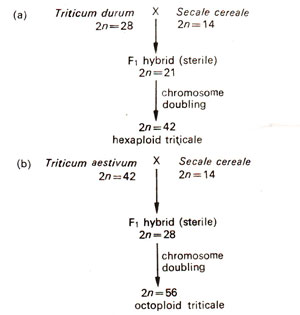
Triticale is the first man made crop, in so far as it resulted as an artificial allopolyploid derived by crossing wheat (Triticum)and rye (Secale). Depending upon whether tetraploid (2n = 4x = 28) or hexaploid (2n = 6x = 42) wheat is utilized for the synthesis, one would get hexaploid triticale (2n = 6x = 42) or octoploid triticale (2n = 8x = 56) respectively. In each case, only diploid rye (2n = 2x = 14) was used. The derivations of triticales are shown in Figure 20.16. In a similar manner, tetraploid triticales (2n = 28) and decaploid triticales (2n = 70) were also produced, but they were not of much significance. In decaploid triticales, tetraploid rye instead of diploid rye was utilized. Tetraploid triticales were produced indirectly through crosses between 6x triticales and 2x rye at some places. However, at present only the hexaploid triticales (2x = 42) are considered to have the potential of becoming a new crop, although in China, octoploid triticales are also being cultivated.
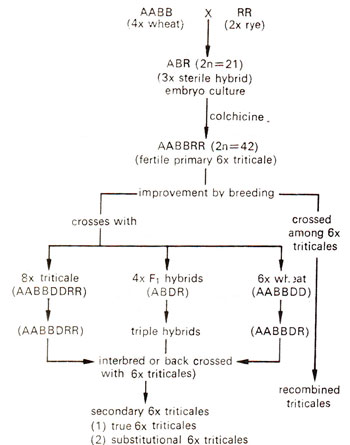 The hexaploid triticales, which are initially produced as amphiploids are called primary triticales. These primary triticales have many drawbacks like shrivelled grains, meiotic instability, poor yield and preharvest sprouting. For further improvement, these primary triticales are regularly intercrossed among themselves and also with either hexaploid wheat or with octoploid triticales followed by backcrosses with hexaploid triticales. General methods of improvement of these triticales are shown in Figure 20.17. It wili be seen that the improved triticales can be either recombined triticales or secondary triticales, the latter resulting due to crosses with 6x wheat or 8x triticales. It is these secondary triticales which are being released as cuitivars in different parts of the world. A list of some of these released triticale cultivars is given in Table 20.5. It will be seen that in India also in 1982 a cultivar. TL 419 was released by Punjab Agricultural University (PAU), Ludhiana, on a regional basis. During the last several years, after the release of TL 419, even better strains giving yields higher than TL 419 could be produced and will be released soon.
The hexaploid triticales, which are initially produced as amphiploids are called primary triticales. These primary triticales have many drawbacks like shrivelled grains, meiotic instability, poor yield and preharvest sprouting. For further improvement, these primary triticales are regularly intercrossed among themselves and also with either hexaploid wheat or with octoploid triticales followed by backcrosses with hexaploid triticales. General methods of improvement of these triticales are shown in Figure 20.17. It wili be seen that the improved triticales can be either recombined triticales or secondary triticales, the latter resulting due to crosses with 6x wheat or 8x triticales. It is these secondary triticales which are being released as cuitivars in different parts of the world. A list of some of these released triticale cultivars is given in Table 20.5. It will be seen that in India also in 1982 a cultivar. TL 419 was released by Punjab Agricultural University (PAU), Ludhiana, on a regional basis. During the last several years, after the release of TL 419, even better strains giving yields higher than TL 419 could be produced and will be released soon.

A more detailed iist of more than 150 triticales was published by CIMMYT (International Centre for Wheat and Maize Improvement) in 1986 (Spring Triticale, Names, Pedigrees, Origins : compiled by O.S. Abdalla et al.)
Segmental allopolyploids.
In some allopolyploids, the different genomes which are present are not quite different from one another. Consequently, in these polyploids chromosomes from different genomes do pair together to some extent and multivalents are formed. This mearts that segments of chromosomes and not the whole chromosomes are homologous. Therefore, such allopolyploids are called segmenta! allopolyploids according to Stebbins (1943, 1950). These segmental allopolyploids are intermediate between autopolyploids and allopolyploids and can be identified by their meiotic behaviour.
It is also believed that most of the naturally occurring polyploids are neither true autopolyploids nor true allopolyploids but are rather segmental allopolyploids. Our common hexaploid bread wheat is also regarded to be a segmental ailopolyploid, because the three diploid genomes (A, B and D) are related (homoeologous) to each other.
(a) Origin and production of autopolyploids.
The autopolyploids may occur in nature or may be artificially produced. When they are found in nature, their autopolyploid nature is inferred mainly by their meiotic behaviour. One of the very common examples of natural autopolyploidy relevant to Northern India, pertains to common 'doob' grass (Cynodon dactylon). In U.P. and Bihar, common 'doob' grass was found to be an autotriploid as inferred from its meiotic behaviour (Gupta and Srivastava, 1970).It is perhaps successful due to efficient vegetative reproduction, because, as will be seen, autoeupolyploids are normally triploids and set no seeds. Autotriploids are also known in watermelons, sugarbeet, tomato, grapes and banana, although in several of these cases the polyploids have been artificially produced. Similarly autotetraploids are known in rye (Secale cereale), corn (Zea mays), red clover (Trifolium pratense), berseem (Trifolium alexandrium), marigolds (Tagetes), snapdragons (Antirrhinum), Phlox, grapes, apples, etc. In Oenothera lamarckiana, an American plant, a giant mutant described by Hugo de Vries was later discovered to be an autotetraploid.
In many cases listed above, autopolyploidy is artificially induced. Since polyploids are normally larger and more vigorous, their role in crop improvement has been realized and techniques developed for artificial induction of polyploidy. Polyploidy is mainly induced by treatment with aqueous solution of a drug called colchicine. This drug has the property of arresting and breaking the spindle so that a cell division without cell wall formation may be affected leading to doubling of chromosome number. The concentration of aqueous solution of colchicine may vary from 0.01 % to 0.50% and the treatment may be given in one of the following manners, (i) Seed treatment may be mainly given by soaking seeds for different durations in. aqueous solution of colchicine. (ii) Injections of colchicine solution may also be given at seeding stage so as to inject solution into cortex tissue with the help of a hypodermic needle, (iii) Axiliary bud treatment is also effective. Since bud is meristematically active, placing cotton soaked in colchicine on the bud and continuous dropping of solution on the bud leads to induction of polyploidy in the branch arising from the treated bud. (iv) Shoot apex treatment is brought about just like bud treatment and is fairly effective, but the shoot apex should come in direct contact of the solution. In order to facilitate this, young leaves covering the shoot apex may be removed.
One of the important effects of polyploidy is to produce 'gigantism'. The autopolyploids may be normally larger in size. Sometimes plants may be smaller than diploids, but leaves, flowers and the cells themselves may be bigger in size. Some of the important effects of polyploidy are a's follows : (i) With increase in cell size, water content increases leading to decrease in osmotic pressure. This results into loss of resistance against frost, etc. (ii) Growth rate decreases due to slower rate of cell division; this leads to a decrease in auxin supply and a decrease in respiration, (iii) In autopolyploids, time of blooming is delayed and prolonged due to slow growth rate, (iv) At higher ploidy level (autooctoploids or higher), the adverse effects are highly pronounced and lead to death of plants.
(d) Cytology of autopolyploids.
In an autopolyploid, there will be more than two sets of homologous chromosomes. This leads to formation of multivalents instead of bivalents as found in diploids. An important difference exists even between autotriploids and autotetraploids, because while in the latter normal disjunction is possible giving rise to diploid gametes, in triploids it is not possible. In an autotriploid, there are three sets of homologous chromosomes. If these three sets are normally paired, trivalents can not disjoin normally and will either disjoin 2 : 1 chromosomes to two poles or will disjoin 1 : 1 leaving one chromosome as a laggard. The number of chromosomes in gametes of triploid organism, therefore, will vary from n to 2n. Most of these gametes are unbalanced leading to high degree of sterility.
(e) Genetics of autopolyploids (trisomic and tetrasomic inheritance).
The segregation pattern in polyploids is quite different than what we find in diploids. In a polyploid there are more than one kind of heterozygotes, because there are more than two homologous chromosomes. The different possible genotypes and expected ratios in autotetraploids are given in Table 20.4. Similar ratios for triplqids are given in Table 20.1. The ratios are based on the assumption that either the gene is close to centromere leading to what we call chromosome segregation or the gene is away from centromere so that crossing over between gene and centromere will take place and ratios will be modified. In the latter case it will be called chromatid segregation, because segregation is taking place not at chromosome level, but at chromatid level due to crossing over.
(f) Uses of induced polyploidy
Since fertility level and seed set are low in induced polyploids, their utility in improvement of crop plants will depend on two major considerations, namely : (i) what is the commercial product, because if seed is not the commercial product low seed set will not be a handicap, and (ii) whether or not fertility level is important. Keeping these requirements in mind, seedless fruits can be produced by using triploids as in case of seedless watermelons produced by H. Kihara (who died in 1986) in Japan. These triploids were obtained from seeds raised by a cross tetraploid x diploid, the tetraploids being raised from diploids by colchicine treatment. This method was also used for obtaining triploids in sugarbeet, tomato and grapes.

Fig. 20.9. Derivation of a tetraploid amphidiploid from two diploid species.

Fig. 20.10. Artificial synthesis of Raphanobrassica.
Polyploidy may also result from doubling of chromosome number in a F1 hybrid which is derived from two distinctly different species. This will bring two different sets of chromosomes in F1 hybrid. The number of chromosomes in each of these two sets may differ. Let A represent a set of chromosomes (genome) in species X, and let B represent another genome in a species Y. The F1 will then have one A genome and another B genome. The doubling of chromosomes in this F1 hybrid (AB) will give rise to a tetraploid with two A and two B genomes (Fig. 20.9). Such a polyploid is called an allopolyploid or amphidiploid.
Raphanobrassica is a classical example of allopolyploidy. In 1927, G.D. Karpechenko, a Russian geneticist, reported a cross between Raphanus sativus (2n = 18) and Brassica oleracea (2n = 8) to produce F1hybrid which was completely sterile. This sterility was due to lack of chromosome pairing, since there is no homology between genomes from Raphanus sativus and Brassica oleracea. Among these sterile F1 hybrids, Karpechenko found certain fertile plants. On cytological examination these fertile plants were found to have 2n = 36 chromosomes, which showed normal pairing into 18 bivalents (Fig. 20.10).
Common cultivated wheat is another important example of allopolyploidy, although its allopolyploid nature has now been questioned. There are three different chromosome numbers in the genus Triticum, namely 2n = 14, 2n = 28 and 2n - 42. The common wheat is hexaploid with 2n = 42, and is derived from three diploid species : (i) AA = Triticum aegilopoides (2n = 14), (ii) BB = Aegilops speltoides ? (2n = 14) (in the past evidence was made available, showing that Ae. speltoides may not be the donor of B genome; it is also believed that the donor of B genome may never be discovered) and (iii) DD = Aegilops squarrosq (2n = 14). The hexaploid wheat, therefore, is designated as AABBDD, the tetraploid (2n = 28) as AABB and diploid (2n = 14) as AA. There is, however, evidence available now which suggests that A, B and D genomes from three diploid species mentioned above are not much different from one another, so that it is believed that the three diploid progenitors of common hexaploid wheat were derived from a common ancestor. For this reason, the common hexaploid wheat is now considered an autopolyploid rather than an allopolyploid. At best, it may be a segmental allopolyploid.
Certain allopolyploids were artificially produced in order to find out the origin of naturally occurring allopolyploids. Common hexaploid wheat and tetraploid cotton furnish two such examples. These two and few other examples will be briefly discussed.

Fig. 20.11. Artificial synthesis of hexaploid wheat.

Fig. 20.12. Artificial synthesis of New World cotton.

Fig. 20.13. Probable origin of Spartina townsendii.
(2) Gossypium hirsutum, popularly known as upland cotton, is another interesting example of amphidiploidy. Old World cotton has 13 pairs of large chromosomes, while American or upland cotton has 13 pairs of smaller chromosomes. The New World cotton, the cultivated long-staple type has 26 paris of chromosomes, 13 large and 13 small. J.O. Beasley crossed the American and Old World cottons and doubled the chromosome number in the F1 hybrids (Fig. 20.12). The amphidiploid thus produced resembled the cultivated New World cotton and when crossed with it gave fertile F1 hybrids. The evidence presented in Figure 20.12, thus showed that New World cotton, the cultivated Gossypium hirsutum (tetraploid) originated from two diploid species, namely G. herbaceum (2n = 26) and G. raimondii (2n = 26).

Fig. 20.14. Probable origin of Primula kewensis.

Fig. 20.15. Probable origin of tetraploid of Galeopsis tetrahit.
(5) Galeopsis tetrahit (2n = 32) could be synthesized (Fig. 20.15) from crosses involving diploid species G. pubescens (2n = 16) and G. speciosa (2n = 16). However, A. Muntzing, who performed these experiments suggested that G. tetrahit is not a strict amphidiploid, but has a tendency towards autopolyploidy.
(c) Triticale (X Triticosecale), a new man made cereal.
In recent years, considerable emphasis has been laid on the possibility of utilizing a new man made cereal known as triticale, on a commercial scale. It is already grown in an estimated area of more than one million hectares and research at several places all over the world is in progress to improve this man made crop. Work done in this crop has been reviewed in detail (Muntzing, 1979, Gupta and Priyadarshan, 1982; Gupta and Reddy, 1991). Several hundred cultivars of triticale have been released during the last 10 years and more cultivars of this crop are being released every year. The area under its cultivation is also fast increasing (for more details consult, Gupta, 1984, 1985, 1987; Gupta and Reddy, 1991).

Fig. 20.17. Methods of improvement of primary triticales to obtain secondary and recombined triticales (from Gupta, 1984).

Fig. 20.17. Methods of improvement of primary triticales to obtain secondary and recombined triticales (from Gupta, 1984).
A more detailed iist of more than 150 triticales was published by CIMMYT (International Centre for Wheat and Maize Improvement) in 1986 (Spring Triticale, Names, Pedigrees, Origins : compiled by O.S. Abdalla et al.)
In some allopolyploids, the different genomes which are present are not quite different from one another. Consequently, in these polyploids chromosomes from different genomes do pair together to some extent and multivalents are formed. This mearts that segments of chromosomes and not the whole chromosomes are homologous. Therefore, such allopolyploids are called segmenta! allopolyploids according to Stebbins (1943, 1950). These segmental allopolyploids are intermediate between autopolyploids and allopolyploids and can be identified by their meiotic behaviour.
It is also believed that most of the naturally occurring polyploids are neither true autopolyploids nor true allopolyploids but are rather segmental allopolyploids. Our common hexaploid bread wheat is also regarded to be a segmental ailopolyploid, because the three diploid genomes (A, B and D) are related (homoeologous) to each other.