Subunit of chromatin - the nucleosome
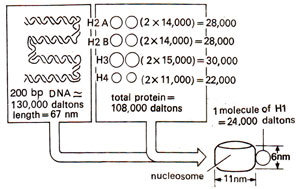
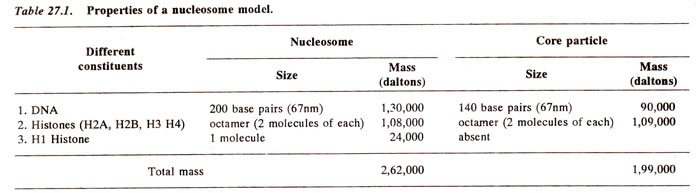
Fig. 27.3. Different constituents (DNA and histone proteins) of a nucleosome and their properties.
Fig. 27.4. Original proposal for nucleosome repeating units, with DNA wound on a series of beads. Actual shape of histone complex or the path of DNA was not known initially (redrawn from Sc. Amer., Vol. 244, 1981).
On the basis of X-ray diffraction, electron microscopy, nuclease digestion, and chemical cross linking, a model for nucleosome structure was proposed. Properties of this nucleosome model are presented in Table 27.1 and Figure 27.3. The shape of nucleosome, arrangement of histones including HI were also studied. The diameter of nucleosome is 11 nm and its height is 6 nm. The length of DNA around it is 70 nm (~ 67 nm DNA = approx. 200 base pairs, because 34 Å = 3.4 nm = 10 base pairs), which is equivalent to 200 base pairs. The length of DNA varies not only in different, organisms (from 154 bp in a fungus to as high as 260 bp in a sea urchin sperm), but also in different tissues or even in different regions within the same cell of an organism. The length of DNA, however, suggested that DNA should be coiled around nucleosome core particle. Enzyme DNAase I also led to cleavage of DNA suggesting that DNA lies on the surface of nucleosome.

Fig. 27.3. Different constituents (DNA and histone proteins) of a nucleosome and their properties.

Fig. 27.4. Original proposal for nucleosome repeating units, with DNA wound on a series of beads. Actual shape of histone complex or the path of DNA was not known initially (redrawn from Sc. Amer., Vol. 244, 1981).
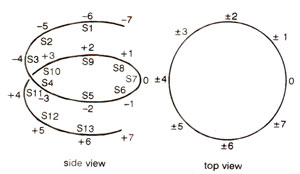
Fig. 27.5. Two numbering schemes dividing core particle DNA into segments, 10 bp apart; SI to S13 are cleavage sites for DNAase I and DNAase II.
The structural periodicity of DNA coiled around the histone octamer, was examined by treatment with enzymes DNAase I and DNAase II. Both of these enzymes make single-strand nicks in DNA, so that double stranded DNA structure is not disturbed. But on denaturation following nicking, single stranded DNA fragments are released.

Fig. 27.5. Two numbering schemes dividing core particle DNA into segments, 10 bp apart; SI to S13 are cleavage sites for DNAase I and DNAase II.
The above results differ from cleavage of DNA immobilized on a flat surface, where the cutting periodicity reflects structural periodicity (10.5 bp per turn). A variation in cutting periodicity in core DNA of nucleosome (10.0 bp at ends; 10.7 bp in the middle) means that there is variation in structural periodicity of core DNA; in the middle, this is close to the value of DNA in solution, but at the ends it is less tightly screwed.
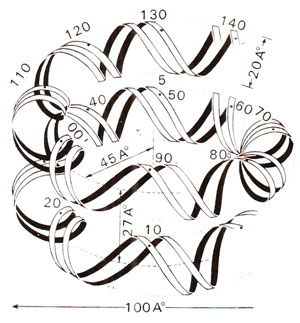
Fig. 27.6. Path of DNA superhelix on a nucleosome core. There are about 80 nucleotide pairs of DNA per turn, one and three quarter turns of DNA are wrapped on histone complex (redrawn from Sci. Amer., Vol. 244, 1981).
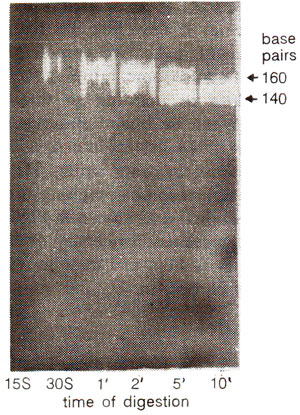
Fig. 27.7. Reduction in the length of nucleosome DNA by nuclease digestion in time (15 seconds to 10 minutes).
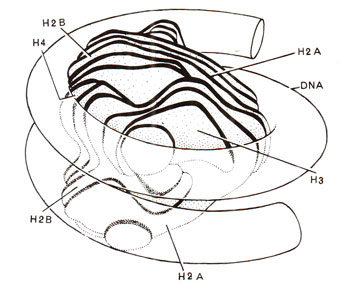
Fig. 27.8. Model of nucleosome core with DNA superhelix wound on histone octamer, which has, a more or less, continuous helical ramp, on which DNA (146 base pairs) is wound (redrawn from Sci. Amer., Vol. 244, 1981).
When nuclease treatment was prolonged beyond the cleavage between nucleosomes, the DNA was removed from both the free ends yielding particles containing DNA of only 146 (or sometimes 140) nucleotide pairs instead of 200 nucleotide pairs (Fig. 27.6). This reduced form of nucleosome is called core particle. Due to nuclease treatment, the length of the DNA is reduced in discrete steps as shown in Figure 27.7. Adjacent core particles, each with core DNA, are joined with each other through DNA segment called linker DNA, which is removed during prolonged digestion. The core DNA has an invariant length of 146 bp, but the linker DNA can vary from 8 bp to 114 bp, thus accounting for variation in DNA length of nucleosomes (154-260 bp).

Fig. 27.6. Path of DNA superhelix on a nucleosome core. There are about 80 nucleotide pairs of DNA per turn, one and three quarter turns of DNA are wrapped on histone complex (redrawn from Sci. Amer., Vol. 244, 1981).

Fig. 27.7. Reduction in the length of nucleosome DNA by nuclease digestion in time (15 seconds to 10 minutes).
The core particles could be obtained in crystalline form for a study of X-ray crystallography to reveal the three dimensional structure. Electron microscopy and X-ray diffraction patterns also revealed that nucleosome core particle is neither spherical, nor perfectly flat, but is wedge shaped, about 1] nm in diameter and 6 nm high. The study of crystals suggested that DNA double helix is coiled into a large helix or superhelix, by making two turns around the histones in the middle of the particle. The two turns of the DNA double helix will almost touch, since DNA is 2 nm thick and the particle is only 6 nm thick. From the structure of DNA and the diameter of core particle, it has been estimated that two superhelical turns would have about 160 base pairs (80 bp per turn). Since length of DNA in a core particle is only 146 pairs, it will occupy only one and three-quarter turns. The particle, therefore, will be thinner on one side, accounting for the wedge shape. It was also shown that histone octamer itself is also bipartite and wedge-shaped providing for a short helical ramp on which the DNA is wound (Fig. 27.8).

Fig. 27.8. Model of nucleosome core with DNA superhelix wound on histone octamer, which has, a more or less, continuous helical ramp, on which DNA (146 base pairs) is wound (redrawn from Sci. Amer., Vol. 244, 1981).