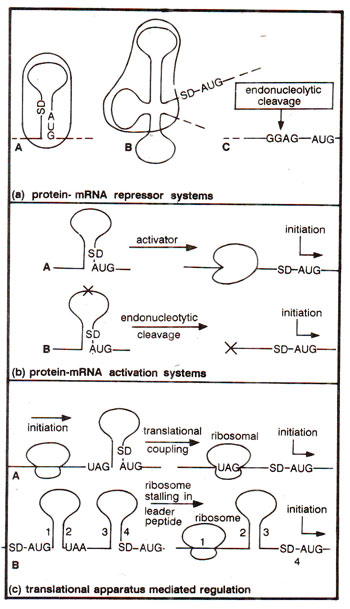
Fig. 35.26. Some models of translational regulation in prokaryotes (repression and activation; see text for details).
The number of regulatory circuits identified at the translational level increased steadily during 1980s and 1990s. Besides, the mechanisms of regulation discussed above, control at the. translational level is also achieved by formation of secondary and tertiary structures, which either inhibit or facilitate-translation. These secondary or tertiary structures are dependent on intramolecular base pairing in mRNA, and can be variously modified due to effect of molecules acting on specific target sites. The regulatory circuits at the translational level may either involve repression or activation.
Repression of translation. Atleast three different situations of repression of translation have been proposed (Fig. 35.26a); (i) A repressor-effector molecule (e.g. phage R17 coat protein) may recognize and bind to a specific sequence or to a specific secondary structure (involving SD region and AUG codon), thus blocking initiation of translation, through blocking of the ribosomal binding region, (ii) A repressor effector molecule may bind to an operator (not involving SD region and AUG codon), thus stabilizing an inhibitory mRNA secondary structure (e.g. threonyl-tRNA synthetase; phage T4 RegA protein, which recognizes TIR sequence domains of a number of T4 genes), (iii) An effector molecule (an endonuclease) can inhibit initiation of translation by endonucleolytic cleavage of SD region (e.g. phage T4
motA mRNA SD region).
Activation of Translation. Cases of translational activation have also been reported, even though they are less well understood. The most likely mode ofaction of these positive effectors or activators is through destabilization of inhibitory secondary structures in mRNA either through binding or by endonucleolytic cleavage (Fig.35.26(b);e.g.phageλ and phageT4 RNaseIII). Translation of certain genes is also influenced by the translation of neighbouring genes, a phenomenon described as translational coupling. Under this mechanism, the ribosome terminating the translation on a preceding gene of a polycistronic mRNA, may sometimes release the inhibitory secondary structure found in the mRNA of the following gene (e.g. 6-phosphogluconate dehydrogenase gene and erythromycin methylase gene; see Fig. 35.26c).

Fig. 35.26. Some models of translational regulation in prokaryotes (repression and activation; see text for details).





