Splicing of eukaryotic hnRNA through spliceosomes/snurposomes
The following two features are significant in the introns of eukaryotic nuclear genes (to some extent, mitochondrial group II introns resemble these nuclear introns, although not entirely) : (i) the two ends of an intron do not have complementary base sequences to allow formation, of secondary structures as has been shown for group I introns; (ii) the exon-intron junctions always have the same sequences, so that an intron in RNA transcript starts with GU (5' end or left border) and ends in AG (3' end or right border); this is sometimes described as GU-AG rule. On either side of intron-exon junctions also, there are common sequences described as consensus sequences, although these may not be found in all cases.
Formation of spliceosome/snurposome. Splicing of intron sequences of eukaryotic hnRNA involves formation of spliceosome complex followed by excision of intron sequence through lariat formation. The major components of the splicing machinery, are snRNPs (or 'snurps') consisting of RNAs and proteins. The snRNAs are designated as Ul, U2, U4, U5 and U6. The role of snRNPs in splicing has been studied in vitro. It has been shown that in association with other essential protein factors and pre-mRNA, the snRNPs form a macromolecular complex known as spliceosome (40-60 nm in size).
These spliceosomes have been isolated in relatively pure form and used for a variety of studies, giving information about the mechanism of splicing. The left splicing junction (GU) is recognized by Ul snRNA and the right splicing junction is recognized by U5. U2 snRNA recognizes another consensus sequence, called branch site present within the intron. Two other snRNAs, U4 and U6 are also involved in formation of spliceosome, but their exact role is not known. The different steps involved in RNA splicing through formation of spliceosome are shown in Figure 33.6. The first step involves association of U2 snRNP followed by addition of U5 and U4/U6 as a single complex. By this stage Ul' is bound to 5' splice site, but its presence can not be easily detected. Therefore, either it associates only transiently or it readily dissociates.
 Recently, snRNPs have been detected in brightly staining granules in the nucleoplasm surrounding the lampbrush chromosomes of amphibian oocytes. Three morphologically distinct types of these granules were identified and called A, B and C snurposomes (A = 1-4 μmwith Ul snRNP; B = 1-4 μm with all five snRNPs; C = 1-20 μm with unknown composition). The functions of these snurposomes are not clearly understood but may include storage of snRNPs and splicing of hnRNA.
Recently, snRNPs have been detected in brightly staining granules in the nucleoplasm surrounding the lampbrush chromosomes of amphibian oocytes. Three morphologically distinct types of these granules were identified and called A, B and C snurposomes (A = 1-4 μmwith Ul snRNP; B = 1-4 μm with all five snRNPs; C = 1-20 μm with unknown composition). The functions of these snurposomes are not clearly understood but may include storage of snRNPs and splicing of hnRNA.
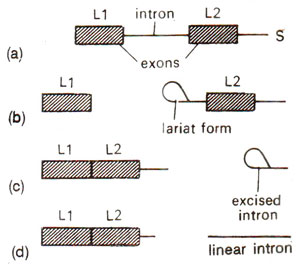
Formation of lariat during splicing. Nuclear splicing, where spliceosome is formed, involves formation of a lariat structure. This occurs in two stages : (i) In the first stage, a cut is made at the left end of the intron, releasing a separate RNA molecule with left exon and a right RNA molecule with intron and the right exons. The two RNA molecules are held together due to components of spliceosome. The left RNA molecule is linear, but the right intron-exon molecule is not linear. The 5' terminus at the left end of intron-exon molecule gets linked by a 5'-2' bond to the A of the sequence CUGAC located - 30 bases upstream of the right end of the intron. This linkage generates a lariat. (ii) In the second stage, cutting at the right splicing junction- releases a free intron in lariat form, and the left exon is Iigated to the right exon. The lariat is debranched to give a linear excised intron, which is rapidly degraded (Fig. 33.7).
Formation of spliceosome/snurposome. Splicing of intron sequences of eukaryotic hnRNA involves formation of spliceosome complex followed by excision of intron sequence through lariat formation. The major components of the splicing machinery, are snRNPs (or 'snurps') consisting of RNAs and proteins. The snRNAs are designated as Ul, U2, U4, U5 and U6. The role of snRNPs in splicing has been studied in vitro. It has been shown that in association with other essential protein factors and pre-mRNA, the snRNPs form a macromolecular complex known as spliceosome (40-60 nm in size).

Fig. 33.7. Formation of lariat type of loop configuration in excised introns in a split gene. LI and L2 are two exons; (a)-(d) are steps in excision.





