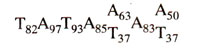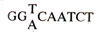As earlier discussed, in eukaryotes there are three RNA polymerases (RNA polymerase I for rRNA; RNA polymerase
II for
hnRNA and RNA polymerase
III for 5S RNA, tRNAs, U6 snRNA, 7SK, EBER2). Promoters for RNA polymerase I could not be initially studied (although recently promoters for this enzyme for rRNA have been studied and their properties described), since all genes for rRNA were similar. Promoters for RNA polymerase
III, on the other hand, had some unusual downstream promoters (to be discussed later). However, for RNA polymerase II, several hundred eukaryotic genes have now been sequenced and their promoters studied revealing some general features in three regions located at start point, centred at sites lying between -25 bp and -100 bp. The least effective of these three regions is the
TATA or
Hogness box (7 bp long) located 20 bp upstream to the start point. The
TATA or
Hogness box has the following sequence (frequency shown by subscripts).
The TATA box is surrounded by G-C rich sequences and is comparable to
Pribnow box of prokaryotes (see
Regulation of Gene Expression 3. A Variety of Mechanisms in Eukaryotes also). Further upstream is another sequence called
CAAT box, which, being necessary for initiation, is conserved in some promoters (β globin gene), but is not necessary in some other genes (thymidine kinase gene of herpes virus). This sequence lies between -70 and -80 base pairs and includes the following.
Another sequence called GC
box (GGGC GG) is found in one or more copies at -60 or -100 bp upstream in any orientation in several genes. It has been shown that CAAT and GC boxes determine the efficiency of transcription, while TATA box aligns RNA polymerase at proper site, with the help of TFIID and other transcription factors. (See later for transcription factors).
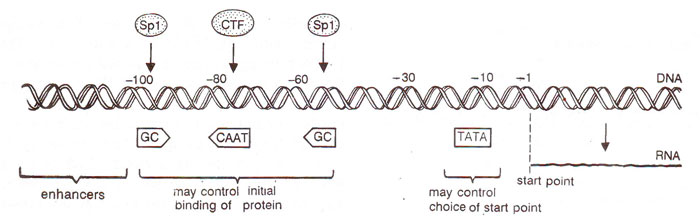
Fig. 32.13. A DNA segment rn a eukaryote showing promoter sites (TATA, GC and CAAT boxes) and the enhancer site.
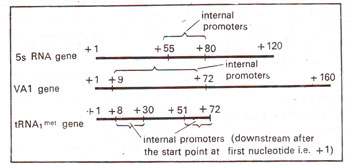
Fig. 32.14. Three different genes transcribed by RNA polymerase III, all containing internal promoters; numbers indicate the number of bases from the start point of the gene.

Fig. 32.15. Binding of TFIIIB to downstream promoters (box A), located at different positions in 5S and tRNA genes.
Eukaryotic promoters often also possess elements located 100 or 200 base pairs upstream, which interact with proteins other than RNA polymerase and thus regulate the activity of promoter. These elements, called
enhancers, also called
upstream activation sites or UAS, can be moved several hundred or even thousands of base pairs upstrearrj,or downstream without alteration of their activities. These enhancers may lead upto a 200 fold increase in transcription rate of an affected gene, hence the name. There are other regulatory elements known as
silencers, which repress gene expression. As shown by
Alexander Johnson (San Francisco), silencers, like enhancers,can function at great distances from genes they repress. They are sites for binding of proteins just like enhancers. The location of different promoters and enhancers is shown in Fig. 32.13. More details about enhancers and their role in gene expression are given in
Regulation of Gene Expression 3. A Variety of Mechanisms in Eukaryotes. (Silencers are found in yeast and repress expression of
HML and
HMR loci involved in switching of mating types; consult
Regulation of Gene Expression 3. A Variety of Mechanisms in Eukaryotes).
In 5SRNA genes transcribed by
RNA polymerase
III, the promoter lies well within the transcription unit, 50 bp downstream rather than upstream from the start point (Fig. 32.14). 5SRNA keeps on being synthesized even when the entire sequence upstream from the gene is removed suggesting that no upstream promoter is involved in transcription. However, deletions of bases downstream from start point, if lie between 55—80 bp lead to complete halt of transcription. Therefore, 50-80bp sequence downstream in 5SRNA gene constitutes the promoter region. The location of downstream promoters for RNA polymerase III .may vary in different genes (Fig. 32.14).
One would be interested in knowing how RNApolymerase III, attached to +55 to +80 bp sequence, can start transcription from the start point. It is believed that the enzyme is big enough to occupy start point and the promoter region simultaneously and thus may start transcribing at the start point without difficulty.

Fig. 32.13. A DNA segment rn a eukaryote showing promoter sites (TATA, GC and CAAT boxes) and the enhancer site.

Fig. 32.14. Three different genes transcribed by RNA polymerase III, all containing internal promoters; numbers indicate the number of bases from the start point of the gene.

Fig. 32.15. Binding of TFIIIB to downstream promoters (box A), located at different positions in 5S and tRNA genes.
The downstream promoter sequences of 5S RNA and tRNA genes have subsequently been delineated more precisely into
box A,
box B and
box C In 5S genes box A and box C are found at +50 to +69 and +80 to +90 respectively. Similarly in tRNA genes, box A and box B are found at +8 to +30 and +51 to +72 respectively. It has also been shown, that box A in 5S genes and tRNA genes, though differs in location, but has similar
; conserved sequences'. It is, therefore, recognised by
the same transcription factor (TFIIIB). These promoter sequences and the transcription factors binding at these sites are shown in Figure 32.15
(see later for more details).
Some RNA polymerase III genes are also known to have upstream promoters like TATA box
(for example U6 snRNA, 7SK, EBER2; genes for
other snRNAs, like U2, are transcribed by RNA polymerase II). Among these genes 7S genes have both types of promoters (downstream and upstream), so that on deletion of box A, upstream promoter is used by RNA polymerase III.