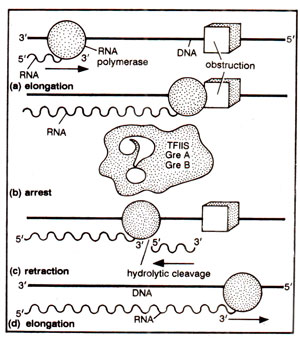
Fig. 32.27. The role of TFIIS in elongation of RNA chain
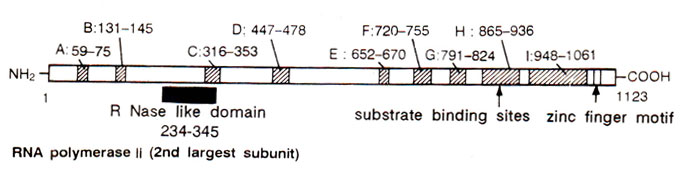
Fig. 32.12. Functional regions of the second largest subunit of RNA polymerase II of Drosophila. Solid bar shows similarity with barnase and other bacterial RNAses; A to I are regions having homology with prokaryotic RNA polymerase.
Certain accessory proteins of transcription, called the
'elongation factors', enhance the overall activity of RNA polymerase II, leading to increase in elongation rate. Atleast two such proteins (transcription factors) are known (i) The transcription factor
TFIIF accelerates RNA chain growth relatively uniformly, in concert with RNA polymerase II or Pol II. (ii) The transcription factor
TFIIS (also called
SII) helps elongation of RNA chain, by relieving the obstructions in the path of such elongation. TFIIS is known to function by first causing hydrolytic cleavage at 3' ends of RNA chains, which are stuck and can not elongate. Thus, RNA polymerase moves backwards (hydrolytic cleavage) under the direction of TFIIS before it moves forward through the block to elongation (Fig. 32.27). It is possible that hydrolytic activity resides in Pol II and is enhanced by TFIIS (second largest subunit of Pol II has regions of homology with bacterial ribonuclease; see Fig. 32.12).
Prokaryotic elongation factors,
Gre A and
Gre B (discovered in 1992-93) also cause hydrolytic cleavage at 3' end of nascent RNA in
E. coli.

Fig. 32.27. The role of TFIIS in elongation of RNA chain

Fig. 32.12. Functional regions of the second largest subunit of RNA polymerase II of Drosophila. Solid bar shows similarity with barnase and other bacterial RNAses; A to I are regions having homology with prokaryotic RNA polymerase.
Termination of RNA synthesis in eukaryotes AAUAAA sequence and 'snurp' in post-transcriptional cleavage. In eukaryotes, the actual termination of RNA polymerase II activity during transcription may take place through termination sites similar to those found in prokaryotes (the nature of individual termination sites in not known). But these termination sites are believed to be present away (sometimes upto one kilobase away) from the site of the 3' end of mRNA. Obviously 3' end of mRNA will be generated due to post-transcriptional cleavage. This cleavage, at the end, is believed to be achieved by what is popularly called
'snurp' (small nuclear RNA-protein complex).
'Snurp' used for post-transcription cleavage has not been identified so far but is believed to be certainly different than the U1
snurp, which is believed to be involved in intron splicing in split genes (see
Expression of Gene : Protein Synthesis 3. RNA Processing (RNA Splicing, RNA Editing and Ribozymes)).
Moreover, a sequence 5'AAUAAA 3' has been found just on the 5' side of poly(A) addition site in several eukaryotic mRNAs (e.g. αand βglobins in mouse and ovalbumin in chicken).We will notice in the next topic that poly(A) tail is added to 3' end of eukaryotic mRNA after processing of precursor mRNA. The sequence 5'AAUAAA3' in mRNA 3' end seems to be common in eukaryotic mRNA and mutations in this sequence cause elongation of mRNA. This will suggest that this sequence contains the signal for endonucleolytic post-transcriptional cleavage. This sequence, therefore, is not involved in the termination of the synthesis of mRNA, but helps in generating 3' end later through endonuclease cleavage, in which
snurp helps in an unknown manner.








