It has been shown that in both chloroplasts and mitochondria, following components of translation apparatus are found : (i) ribosomes specific to organelle, (ii) tRNAs specific to organelle, (iii) other factors for translation (little is known about these factors). Some of these components may be synthesized outside these organelles, and then transported into organelles.
It has been shown that the translation apparatus in chloroplasts and mitochondria differ from that in cytoplasm in eukaryotes in the following respects, (i) Ribosomes in these organelles are smaller in size (70S) than those in cytoplasm (80S), (ii) The tRNAs are specific and differ, the number of tRNAs in mitochondria being 22 as against 55 in cytoplasm, (iii) Initiation of translation takes place by formyl-methionyl tRNA both in chloroplasts and mitochondria, although no formylation takes place in cytoplasm, (iv) Translation in chloroplasts and mitochondria can be inhibited by chloramphenicol, as in bacteria since the 70S ribosomes are sensitive to chloramphenicol and not to cycloheximide; on the other hand, the translation in cytoplasm is inhibited by cycloheximide, since 80S ribosomes are sensitive to cycloheximide.
There are other antibiotics like spectinomycin, lincomycin and erythromycin which also inhibit translation in bacteria and in all organelles of eukaryotes. These antibiotics help to stop the translation preferentially either (i) in cytoplasm (by cycloheximide) thus permitting protein Synthesis only in chloroplasts or mitochondria or (ii) in organelles (by chloramphenicol) thus permitting protein synthesis only in the cytoplasm.
Studies on the synthesis of
'chlorophyll-binding proteins', which play a major role in the primary reactions of photosynthesis have also provided new information about chloroplast translation and its regulation. In many plant species, in the absence of light the mRNA of chlorophyll binding proteins, including those encoded by chloroplast genome, remain associated with thylakoid bound polysomes, but are not translated. Transfer of seedlings to light induces synthesis of proteins, although no increase in transcription has been observed. This suggests that post-transcriptional processes play a key role in light induced chloroplast gene expression.
It has also been shown that the polycistronic mRNA encoded by
the psbB operon in maize, need not be cleaved into tri-, di- or monocistronic forms for successful translation (
psbB operon =
psbB +
psbH +
petB + petD)
. This suggests that plastid ribosomes can bind directly to internal initiation regions for initiation of translation as in prokaryotes. However, RNA processing does take place in chloroplasts and mitochondria, and the transcripts may differ in their translatability.
There are also nuclear genes, whose products are essential for translation of specific genes. Several of these nuclear encoded factors interact with the 5' untranslated region of chloroplast massages, but little is known about the identity and precise function of these factors. These products may be translational activators of the type described in
Regulation of Gene Expression 1. Operon Circuits in Bacteria and other Prokaryotes and
Regulation of Gene Expression 3. A Variety of Mechanisms in Eukaryotes for prokaryotic and eukaryotic genes respectively.
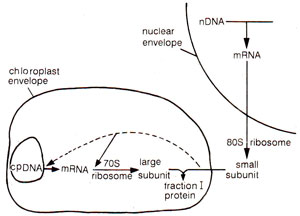
Fig. 34.14. A model of co-operation between nuclear DNA and chloroplast DNA in pea as studied by R.J. Ellis and his colleagues (redrawn from Nature Vol. 254, 1975).
Protein synthesis in isolated chloroplasts of pea was studied by R.J. Ellis of U.K. The proteins which could be synthesized in isolated chloroplasts included (i) large subunit of Fraction I protein, (ii) five unidentified proteins of the internal lamellar system, and (iii) two or three unidentified polypeptides of the envelope. These are only few of a large number of proteins found in chloroplasts. It has also been demonstrated that while the large subunit of Fraction I protein is synthesized under the influence of chloroplast DNA the small subunit is synthesized in the cytoplasm under the influence of nuclear DNA. Small subunit is then transported to the chloroplast where it associates with large subunit to give rise to Fraction I protein (Fig. 34.14).
Proteins synthesized in mitochondria have also been identified by inhibiting protein synthesis in cytoplasm by cycloheximide. Although, some proteins are synthesized in mitochondria, not all proteins present in mitochondria are synthesized there. There are some mitochondrial proteins which are synthesized in cytoplasm and then transported. There are still others having dual origin, some polypeptides having cytoplasmic origin and others having mitochondrial origin. Some proteins, which are known to be synthesized in mitochondria include the following : (i) Cytochrome oxidase contains seven different kinds of polypeptides, of which three are synthesized on mitochondrial ribosomes, the remaining four being synthesized on cytoplasmic ribosomes. (ii) Adenosine triphosphatase (ATPase) is an important component of mitochondrial membrane, which plays an important role in coupling respiration to ATP formation. There are other polypeptides present in this complex and only two are definitely known to be synthesized on mitochondrial ribosomes. (iii) In mitochondrial ribosomes, although ribosomal proteins are known to be synthesized outside mitochondria and under the influence of nuclear DNA, but rRNA is transcribed from mtDNA, since both 21S rRNA and 15S rRNA (in yeast mitochondria, 21S rRNA and 15S rRNA occur instead 23S and 16S rRNAs;) hybridize with specific regions of mtDNA.

Fig. 34.14. A model of co-operation between nuclear DNA and chloroplast DNA in pea as studied by R.J. Ellis and his colleagues (redrawn from Nature Vol. 254, 1975).





