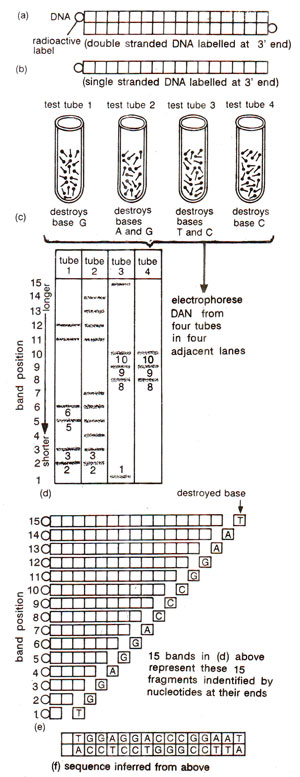
Maxam and Gilbert's chemical degradation method

Fig. 41.7. Dideoxynucleotide and the strategy involved in Sanger's chain termination method for sequencing of DNA.

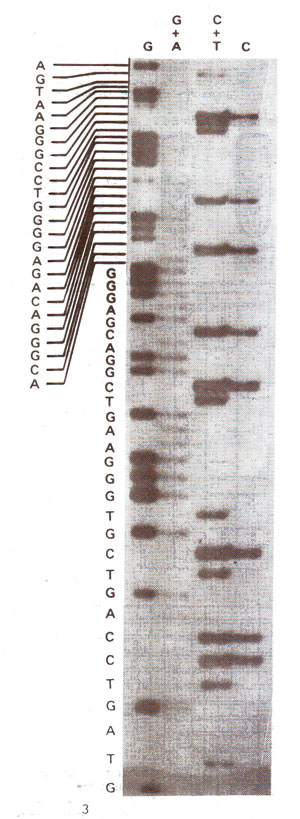
Fig. 41.8. An autoradiograph of Sanger's sequencing gel and the nucleotide sequence read from it.
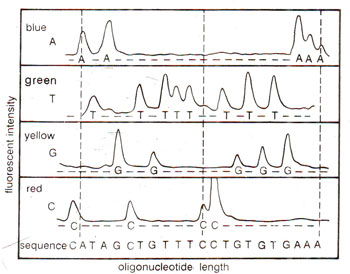
Fig. 41.9. Fluorescence detection of oligonucleotide fragments produced by Sanger's dideoxy chain termination method (redrawn from Nature, 324 : p.674, 1986).
These dideoxy nucleotides are used as triphosphates (ddNTP) and can be incorporated in a growing chain, but they terminate synthesis, since they can not form a phosphodiester bond with next incoming nucleotide triphosphate (dNTP). Following steps are involved in Sanger's dideoxy method for DNA sequencing,
(i) Four reaction tubes are set up, each containing single stranded DNA sample (cloned in M13 phage) to be sequenced, all four dNTPs (radioactively labelled), an oligonucleotide primer, and an enzyme for DNA synthesis (DNA polymerase I). Each tube also contains a small amount (much smaller amount relative to four dNTPs) of one of the four ddNTP, so that four tubes have each a different ddNTP, bringing' about termination at a specific base-adenine (A), cytosine (C), guanine (G) and thymine (T).
(ii) The fragments generated by random incorporation of ddNTP leading to termination of reaction are then separated by electrophoresis on a high resolution polyacrylamide gel. this is done for all the four reaction mixtures on adjoining lanes in the gel.
(iii) The gel is used for autoradiography so that the position of different bands in each lane can be visualized,
(iv) The bands on autoradiogram can be used for getting, the DNA sequence as shown in Figure 14.8.
A variant of the above dideoxy method was later developed, which has allowed the production of automatic sequencers. In this new approach, a different fluorescent dye is tagged to the oligonucleotide primer in each of the four reaction tubes (blue for A, red for C, etc.). The four reaction mixtures are pooled and electrophoresed together in a single polyacrylamide gel tube. A high sensitivity fluorescence detector is placed near the bottom of the tube, which measures the amount of each fluorphore as a function of time. The sequence is determined from the temporal order of peaks corresponding to four different dyes (Fig. 41.9).

Fig. 41.7. Dideoxynucleotide and the strategy involved in Sanger's chain termination method for sequencing of DNA.

Fig. 41.8. An autoradiograph of Sanger's sequencing gel and the nucleotide sequence read from it.

Fig. 41.9. Fluorescence detection of oligonucleotide fragments produced by Sanger's dideoxy chain termination method (redrawn from Nature, 324 : p.674, 1986).