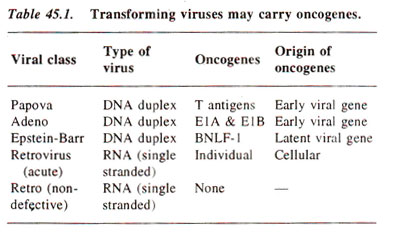
Certain tumour viruses (including both DNA and RNA viruses) carry genes, which confer on them the ability to convert host cells to a tumorigenic state. These genes are called oncogenes (Table 45.1). Such DNA and RNA viruses differ in some important characteristics which will be briefly described : (a)
DNA viruses. The DNA tumour viruses may also grow in culture on a variety of cells, which can be classified into following two types : (i)
'permissive cells', which can be productively infected and (ii)
'non-permissive cells', which are not productively infected, but merely get transformed (Fig. 45.1). Most oncogenes in these viruses are expressed by alternative splicing (consult
Expression of Gene : Protein Synthesis 3. RNA Processing (RNA Splicing, RNA Editing and Ribozymes)). Unlike RNA viruses, oncogenes in these DNA viruses do not have cellular counterparts (see later), (b)
RNA viruses. RNA tumour viruses may be transformation competent (able to transform the infected cells) or transformation-incompetent (able to replicate, but not transform the infected cells). They may replicate via DNA intermediate or an RNA intermediate and can transfer genetic information horizontally or vertically (Fig. 45.2).
The RNA viruses may be classified in two groups with respect to their tumorigenicity. (i)
Non-defective viruses. These viruses cause tumour formation, not due to an individual oncogene, but due to its ability to activate a cellular gene(s); two classic models are FeLV (Feline leukemia virus; Feline = of cats) and MMTV (mouse mammary tumour virus), (ii)
Acute transforming viruses. These viruses, each possesses an oncogene (absent in the ancestral virus, but derived later from host cell due to a transducing agent), which induces tumour formation
in vivo rather rapidly, and transforms cultured cells
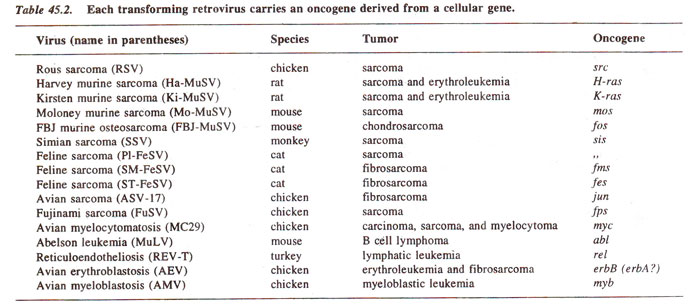
in vitro. These transforming viruses are classified further according to the type of tumour, which is caused (e.g. leukemia, sarcoma, carcinoma, etc.), and the type of tumour is dependent on the oncogene, a virus carries. Oncogenes of some retroviruses are listed in Table 45.2.
These oncogenes are related to some sequences in the cellular genome, called
proto-oncogenes, which are not oncogenic, but can give rise to an oncogene, particularly when captured by a retrovirus. The viral oncogenes and cellular proto-oncogenes are distinguished by using prefixes
v and
c so that an oncogene carried by Rous Sarcoma Virus is called
v-src and the related proto-oncogene in the cellular genome is called
c-src. v-onc is actually derived from m-RNA, so that it contains uninterrupted sequence (or sequence missing at one or both ends), while the corresponding
c-onc may be a split gene with exons and introns. The
mos, ras, sis and
myc genes have been derived from entire
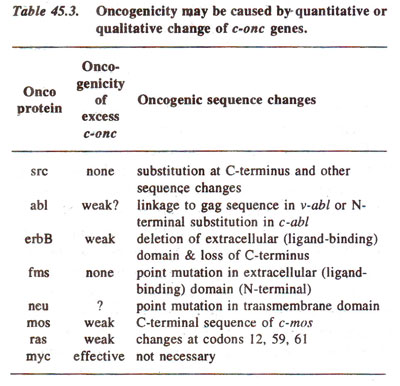
c-onc genes having no missing regions (Table 45.3). A
v-onc sequence is often expressed as a part of a fusion product, where the boundaries can be defined by comparing with the corresponding
c-onc. Several such examples include gab-abl (from
v-abl)
, gag-fps (from
v-fps)
, gag-myc (from
v-myc)and gag-fos (from
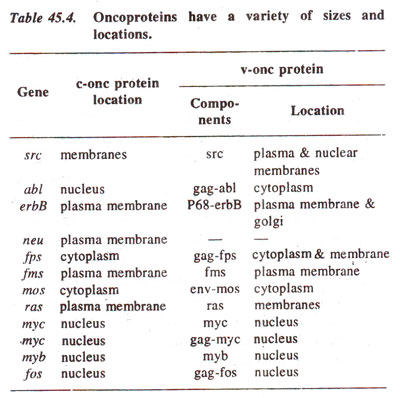
v-fos) (see Table 45.4).
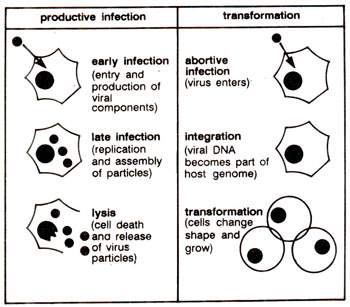
Fig. 45.1. Response of permissive cells and non-permissive cells to virus infection; permissive cells are productively infected by DN A tumour virus, which leads to lysis, but non-permissive cells get transformed through change in their phenotype.
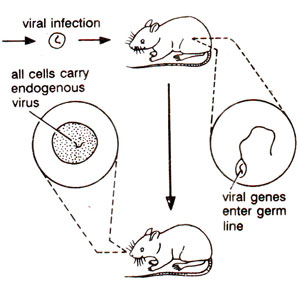
Fig. 45.2. Horizontal and vertical transfer of genetic information from retroviruses; horizontal transfer involves infection of new host, while vertical transfer involves transfer from one generation to the next generation, after integration of viral genome in the germ line of the host.
Atleast 20
c-onc genes have been identified by their representation in retroviruses. Same
c-onc gene may be found in more than one retroviruses and more than one
c-onc gene may be present in different strains of the same retrovirus, although the different
v-onc genes may also be related (derived from different members of same
c-onc gene family^ e.g.
c-ras genes or
c-myc genes).
A wide variety of mechanisms that activate proto-oncogenes (
c-onc)are listed in Table 45.3.
That the
v-onc sequence is actually responsible for oncogenic potential of a transforming virus, has been proved by using following two approaches : (i)
v-onc genes could be mutated to abolish its transforming activity; (ii)
c-onc proto-oncogenes can be altered to become oncogenic. Changes in protein sequence of the product derived from
c-onc, relative to that derived from
v-onc is not always necessary for oncogenicity. In some cases, mere over-expression or altered regulation may lead to oncogenic phenotype.