In the two models for recombination discussed above, it has been shown that heteroduplex DNA regions are produced, which are associated with flanking markers that may or may not have undergone crossing over. The formation of heteroduplex segments is the only plausible means of interaction between DNA duplexes. It also provides an explanation for the phenomenon of gene conversion at a locus, which thus, may or may not be associated with crossing over between the markers flanking this locus undergoing gene conversion. If the region of heteroduplex formation is homozygous at the locus under study, this will not be subjected to any gene conversion, because the strand of a duplex, although derived from non-sister chromatids, are still similar (no mismatch).
But if there is heterozygosity at the locus under study and it falls in the heteroduplex or hybrid DNA region, then there will be mismatch between two strands of a DNA molecule. This may then lead to gene conversion in either of the two directions (
+/a→
+/+ or
a/a)
. This mismatch actually means non-complementary base pairs, so that the heteroduplex DNA segment will be unstable. This mismatch will be recognized by the mismatch repair system in the same manner as thymine dimers, induced by UV light, are recognized by an excision repair system.

Fig. 13.5. Origin of mismatches in heteroduplex DNA during recombination; (a) four chromatids of two paired homologous chromosomes carrying different alleles (+ = GC; m = AT); (b) breakage of single strands, which cross over to join at one another's breakage point; (c) formation of half chromatid chiasma and two stretches of heteroduplex DNA, (carrying mismatches-GT and CA) extending between breakpoints and chiasma.
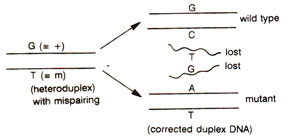
Fig. 13.6. Correction of mismatched nucleotides in heteroduplex DNA to either wild type or mutant type; the mismatched nucleotides produce a distortion, that will be recognized by a repair system (see
Chemistry of the Gene 2. Synthesis, Modification and Repair of DNA for more details on DNA repair).
The above may be illustrated using a hypothetical example, where a heterozygote is
+/m in which + and
m differ by single base pair, their being G-C base pair in + allele and A-T base pair in
m allele at the corresponding position. At meiosis, each of the four chromatids belonging to two paired homologous chromosomes, will have one DNA duplex thus making four DNA duplexes, of which, at a specific point, only two are involved in recombination process. After interaction of DNA molecules, in heteroduplex segments, mismatches may by represented by G-T pair in one duplex and by C-A base pair in the other (Fig. 13.5). If we consider the fate of only one base pair i.e. G-T, this mismatch may be repaired to restore G-C base pair i.e. + allele or may give rise to A-T base pair i.e.
m allele (Fig. 13.6). If no mismatch repair is affected, then the mismatch may be corrected through replication at the first post meiotic mitotic division giving post meiotic segregation. Therefore, the heteroduplex segments may have any one of the following fates shown in Table 13.1 : (i) no mismatch repair or correction, leading to aberrant 4 : 4 segregation with two mixed spore pairs (
mm, m+, m+, ++); this represents post-meiotic segregation; (ii) mismatch repair or correction in one heteroduplex DNA giving 3 : 5 ratio (
mm, mm, m+, ++), or 5 : 3 ratio (
mm, m+, ++, ++); this represents, both gene conversion and post meiotic segregation (pms); (iii) mismatch repair in both heteroduplexes giving a 2 : 6 ratio (
mm, mm, mm, ++) or 6 : 2 ratio (
mm, ++, ++, ++); this represents gene conversion without pms. Other uncommon ratios are also found and can be explained by mismatch repair at one or both heteroduplex segments in either of the two directions.

Fig. 13.5. Origin of mismatches in heteroduplex DNA during recombination; (a) four chromatids of two paired homologous chromosomes carrying different alleles (+ = GC; m = AT); (b) breakage of single strands, which cross over to join at one another's breakage point; (c) formation of half chromatid chiasma and two stretches of heteroduplex DNA, (carrying mismatches-GT and CA) extending between breakpoints and chiasma.

Fig. 13.6. Correction of mismatched nucleotides in heteroduplex DNA to either wild type or mutant type; the mismatched nucleotides produce a distortion, that will be recognized by a repair system (see
Chemistry of the Gene 2. Synthesis, Modification and Repair of DNA for more details on DNA repair).
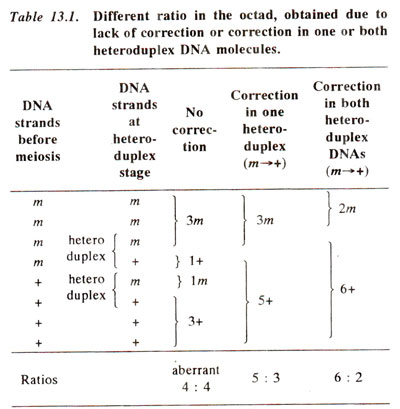
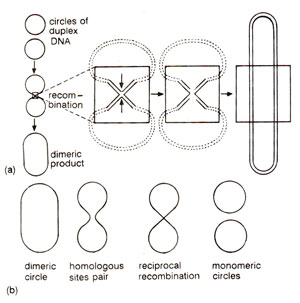
Fig. 13.7. (a) Formation of a dimeric circle from two duplex circles through recombination, involving an intermediate structure resembling a figure of eight (8); (b) formation of two monomeric circles from a dimeric circle by recombination between homologous sequences.

Fig. 13.8. A chi structure with four duplex arms held together at the site of strand exchange (based on an electron micrograph).
Thus the phenomenon of gene conversion provided actual clues for the formulation of hybrid or heteroduplex DNA model for explaining the mechanism of genetic recombination or crossing over at the molecular level. Several features or events involved in the hybrid DNA models have been experimentally demonstrated
in vivo or
in vitro. These include (i) nicking, (ii) strand displacement, (iii) branch migration, (iv) repair synthesis and (v) ligation. Recombination intermediate structures envisaged in Holliday's model and other hybrid DNA models have also been observed under the electron microscope in recombining phages and plasmids. Figure 13.7. shows how two duplex circles (as found in plasmids) may give rise to a dimeric circle through an intermediate structure resembling a figure of eight (8) and how a dimeric circle can give rise to two monomeric recombinant circles. Structures like figure of eight may be isolated and will give rise to chi (χ)structures due to cleavage with a restriction enzyme, that cuts each monomeric circle once. This structure will have four arms held together by a region of heteroduplex DNA at the point of fusion of four arms. Such a chi (χ) structure has actually been observed and one such structure is shown in Figure 13.8.

Fig. 13.7. (a) Formation of a dimeric circle from two duplex circles through recombination, involving an intermediate structure resembling a figure of eight (8); (b) formation of two monomeric circles from a dimeric circle by recombination between homologous sequences.

Fig. 13.8. A chi structure with four duplex arms held together at the site of strand exchange (based on an electron micrograph).













