Genetic investigations on cell division have been conducted on yeasts,
Drosophila and
Aspergillus, but the most important of these studies were conducted on two yeasts, the
budding yeast
(Saccharomyces cerevisiae) and the
fission yeast (Schizosaccharomyces pombe). In both these yeasts, a large collection of cell division cycle
(cdc in fission yeast;
CDC in budding yeast) mutants were isolated, which arrest the cell cycle at specific points described as checkpoints. The interactions
of these mutants were analysed and the products of mutant genes were compared with the macromolecules isolated and studied by those who studied the biochemistry of the cell division (Table 8.1).
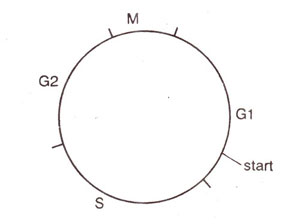
Fig. 8.1. Phases of the cell cycle in the budding yeast.
The G1 phase of the cell cycle has been divided into
pre-start G1, start and
post-start G1 stages (Fig. 8.1). In the cell cycle of budding yeast, 'start' is particularly an important control point, where the mating
pheromones (α factor and
a factor produced by the αand
a haploid cells) cause cell cycle arrest, allowing cells to undergo conjugation. In mammals, passage of the cell cycle through ‘start’ (also called restriction point) requires that (i) the cell attains a minimum size,
so that unless this size is attained the cell cycle will not proceed beyond ‘pre-start G1’; (ii) the cells are not in short supply of nutrients, so that if the cells are starved, they will not pass through 'start', even after attaining the required minimum size; (iii) the cells do not have any mating 'pheromones' and are thus not made to undergo conjugation. Mutants have been isolated, which mimic defects in all these different conditions.

Fig. 8.1. Phases of the cell cycle in the budding yeast.
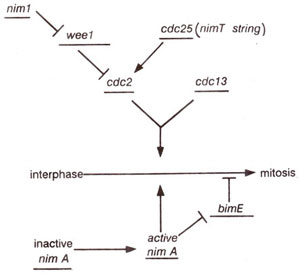
Fig. 8.2. A model for induction of mitosis in the fission yeast. Gene products are shown as stimulating (→) or inhibiting (—|) the activity of other gene products. The cdc2 and cdc13 gene products are shown acting in concert to induce mitosis.
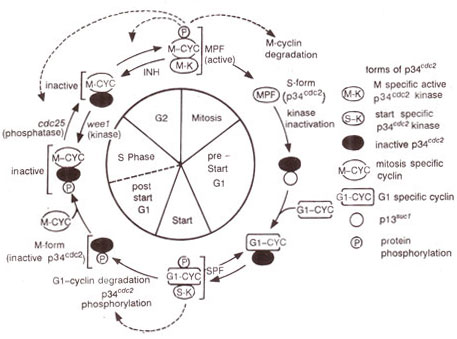
Fig. 8.3. A model of the cycle of cdc2 gene product (p34cdc2) during the cell cycle. (Note that p34cdc2 molecule lacks kinase activity during most of the cell cycle, but becomes active as MPF at mitosis and SPF at start as a result of association with M cyclins and Gl cyclins respectively; active forms can also stimulate their own activation and/or inactivation (→); the dashed line between post start Gl and S phases indicates that no change in p34cdc2 occurs during onset of DNA synthesis.
Genes for passage through Gl and G2 phases Cell division cycle genes (cdc2, cdcl3, cdc25) and weel gene in fission yeast.
A study of mutants suggested that the
cdc 2 gene in fission yeast and
CDC 28 gene in budding yeast are responsible both for passage through start (i.e. transition from Gl to S phase), and for transition from G2 to M phase. Therefore, these two genes are believed to be equivalent to each other. The protein product of these genes and that of their homologues in other organisms is called p34
cdc2 (mol. wt. 34,000 daltons), which is found in all eukaryotic cells. In fission yeast, another gene
cdc 13 (which produces
cyclin or its homologue) is also required for the induction of mitosis and a physical interaction between the products of
cdc 2 and
cdc 13 (p34
cdc2 and cyclin) has been demonstrated. The coordinated activity of
cdc 2 and
cdc 13 is regulated by two additional genes,
cdc 25 (stimulates entry into mitosis), and
wee 1 (inhibits entry into mitosis), which act antagonistically to control and regulate the entry of cell into mitosis. Another gene
nim 1 (see next section) exercises a negative control on
wee 1 (Fig. 8.2). -Both
nim 1 and
wee 1 are believed to produce kinases (kinases cause phosphorylation) and
cdc 25 produces a phosphatase (phosphatases cause dephosphoryla-tion), which removes phosphate from a tyrosine residue(tyr-15) of p34
cdc2 Therefore, these genes act through regulation of protein phosphorylation and dephosphorylation.
An increase in the ratio of activities of two genes, namely that of
cdc 25 to
wee 1, increases the cell size required for entry into mitosis, while a decrease in this ratio leads to decrease in this critical cell size. The overexpression of
cdc 25 in a strain, mutant for
wee 1, causes premature entry into mitosis resulting into lethality. It has also been shown that a
Drosophila gene named
string (required for later embryonic cell cycles) is homologous to
cdc 25.
A model for the pathway that leads to the induction of mitosis has been constructed from genetic analysis of the above mutants and is presented in Figure 8.3.

Fig. 8.2. A model for induction of mitosis in the fission yeast. Gene products are shown as stimulating (→) or inhibiting (—|) the activity of other gene products. The cdc2 and cdc13 gene products are shown acting in concert to induce mitosis.

Fig. 8.3. A model of the cycle of cdc2 gene product (p34cdc2) during the cell cycle. (Note that p34cdc2 molecule lacks kinase activity during most of the cell cycle, but becomes active as MPF at mitosis and SPF at start as a result of association with M cyclins and Gl cyclins respectively; active forms can also stimulate their own activation and/or inactivation (→); the dashed line between post start Gl and S phases indicates that no change in p34cdc2 occurs during onset of DNA synthesis.
‘Never-in-mitosis (nim)' and ‘blocked-in-mitosis (bim)’ genes in Aspergillus. In
Aspergillus nidulans, recently a number of temperature sensitive cell cycle mutants were isolated, which could be classified in two groups : (i)
'never-in-mitosis' (nim) mutants and (ii)
‘blocked-in-mitosis’ (bim) mutants. Several
nim mutants arrest cell cycle in G2 and therefore their normal allele
nim+ must be responsible for functions involved in inducing mitosis. One of these genes (or alleles)
nim T is a homologue of
cdc 25 of fission yeast and
MIH1 gene of budding yeast. The product of these genes is a protein phosphatase that removes phosphate from a tyrosine residue on p34
cdc2. The presence of this phosphate group inhibits protein kinase activity of p34
crfc2 and its removal by a phosphatase (product of
cdc 25) is a rate limiting step for the entry into mitosis.
Another mutant in mitosis gene
nim A also arrests mitosis in G2. When
nim A overexpresses in its mutant form, it induces premature entry into mitosis. Therefore,
nim A
+ should also play an important role in the induction of mitosis. The gene
nim A+ (wild allele of
nim A) encodes a protein kinase called NIM A, whose kinase activity rises as cells enter into mitosis and falls as soon as the mitosis is over. When kinase activities of NIM A and p3
cdc2 were compared in different cell cycle mutants, it could be shown that activations of two protein kinases (p34
cdc2 and NIM A) proceed through independent pathways, although both kinases need to be activated in order (one after the other), so that cells may enter mitosis.

Fig. 8.4. Interactions between p34cdc2 , nim A and bim E and the induction of mitosis (—| indicates inhibitory interactions).
Another temperature sensitive mutant
bim E enters mitosis from any point in the cell cycle, when placed in non-permissive temperature. In double mutants
bim E nim A, the mutation
nim A does not block the ability of mutation
bim E to enter mitosis. It means that in single mutation
nim A, the wild allele
bim E+ actually inhibits entry into mitosis and this inhibitory effect is overcome by the activation of
nimA in its normal form
nim A+, producing the kinase NIM A.
The mitosis spindle is also normal in
bim E, but defective in double mutant
bim E nim A confirming that
bim E mutant overcomes the inhibitory effect of
bim £
+, while
nimA mutant loses the function of phosphorylation performed by NIM A, which may be required for spindle formation and for entry into mitosis. The interactions of p34
cdc2, NIM A and BIM E (or the genes
cdc 2,
nim A, and
bim E) are shown in Figure 8.4.

Fig. 8.4. Interactions between p34cdc2 , nim A and bim E and the induction of mitosis (—| indicates inhibitory interactions).
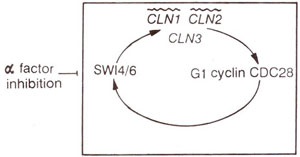
Fig. 8.5. Positive feedback loop involving budding yeast Gl cyclins. Wavy lines showthe genes thought to be activated at the transcriptional level by SW14/6; a factor is an inhibitor of the feedback loop and behaves as a negative growth factor for yeast.
Cyclin genes involved in Gl phase
Cyclin genes in budding yeast. As will be shown later, two types of cyclins are mainly involved in mitotic cell division, G1
cyclins (also called
Cln) and
Mtmitotic) cyclins (also called
cyclin B). Gl cyclins have an essential role in
start, and three cyclin genes
(CLN1, CLN2, CLN3) were originally identified in budding yeast. The messenger RNA and protein levels of CLN 1 and CLN2 undergo periodic variation with the cell cycle, peaking in late Gl, but that of CLN3 is constantly low throughout the cell cycle. The Gl cyclins control the transcription of the CLN1 and CLN2 genes through a positive feedback loop.
In addition two more genes
SW14 and
SW16 (and also
CDC 28) are also required for start dependent transcription of
HO gene coding for an endonuclease responsible for gene-switching at the mating type locus (consult
Regulation of Gene Expression 3. A Variety of Mechanisms in Eukaryotes).
SW14 and
SW16 (SWI stands for switching) are also required for transcription of
CLN1 and
CLN2 (Fig. 8.5); a factor inhibits
SW14 and
SW16 to bring about arrest of cell division to facilitate conjugation (In budding yeast 'a' factor and 'a' factor are two mating type factors). SW14 and SW16 (products of genes
SW14 and
SW16) are components of a transcription factor (SBF) that binds the promoter elements (SCBs = CACG AAAA) responsible for activation of
HO, CLN1 and
CLN2 genes. An analogy has been drawn between components of the above pathway and tumour suppression genes. If these genes are absent, cancer may be caused, and if positive feedback loop is dampened, the cells stop undergoing rapid cell divisions.

Fig. 8.5. Positive feedback loop involving budding yeast Gl cyclins. Wavy lines showthe genes thought to be activated at the transcriptional level by SW14/6; a factor is an inhibitor of the feedback loop and behaves as a negative growth factor for yeast.
Cyclin genes in mammals. In mammals, an oncogene
PRAD1 has been shown to code for a Gl cyclin and is similar to a mouse cyclin gene
CYL1. Four classes of human cyclin genes
(cyclin C,
D, E, F) have also been shown to code for Gl cyclins.
Genes for dependence of mitosis on DNA synthesis
In several organisms including yeast, mammals (tissue culture cells),
Aspergillus, etc. it has been shown that mitosis can be arrested by the inhibitors or by inactivation of replication enzymes due to mutations. In yeast,
RAD 9 gene specifies a component of this control system. A number of temperature sensitive mutants defective in DNA synthesis machinery have been isolated, which do not normally undergo mitosis at the restrictive temperature. These mutations (or cells damaged by irradiation) involve fission yeast genes
cdc 17 (DNA polymerase I),
cdc 9 (DNA polymerase II) and
cdc 2 (DNA polymerase
III), the corresponding genes in budding yeast being
POL1, POL2 and
POL3. However, this dependence of mitosis on DNA synthesis in these mutants is relieved by a complete deficiency of
RAD 9.
In other words, if these
cdc or
POL mutants are combined with
RAD 9 gene defect, then the cells continue into the next cell cycle at the restrictive temperature. It means that
RAD 9 may inhibit mitosis in these mutants and the mutation in
RAD 9 releases this negative control, thus permitting mitosis to be completed in these mutants even when DNA synthesis is interrupted. It is speculated that
RAD 9 gene product may interact with MPF or other components known to play essential roles in the initiation of mitosis.
In budding yeast
(S. cerevisiae), expression of 17 genes is required for DNA synthesis, and their expression is co-regulated due to presence of a common upstream regulatory sequence 5'-ACGCGTNA-3'. This sequence is described as MCB
(Mlu I cell cycle box), since it represents a recognition site for
Mlu I enzyme. Two protein factors, MBF factor and DSC 1 (DNA synthesis) recognize MCB. A gene
cdc 10+ codes for a component of DSC 1, which regulates
cdc22+ (gene for ribonucleotide reductase subunit), and possibly other genes of cell cycle. This
cdc 10+ gene product is believed to be a transcription factor like several other proteins involved in cell cycle.
Gene for dependence of centrosome duplication on DNA synthesis
When DNA synthesis is inhibited in the temperature sensitive mutants, or when
aphidicolin is added in yeast and mammalian cells respectively, centrosome duplication is arrested. In yeast, due.to a mutation
esp 1, although DNA synthesis is blocked leading to inhibition of nuclear division, but centrosome duplication continues leading to accumulation of as many as eight centrosomes within the same cell. This proved the presence of a special gene, which prevents centrosome duplication in the absence of DNA synthesis.
Genes for dependence of mitosis on spindle function
It has been shown that a microtubular spindle is required for proper segregation of the chromosomes, so that an impaired spindle function leads to chromosome missegregation. This is
suggested from the observation that whenever the spindle is disrupted with microtubular poisons, such as
benomyl or
nocodazole, progression through mitosis is blocked. Mutants have also been isolated in the budding yeast
(S. cerevisiae), which complete mitosis and cell division, even when spindle is not functioning properly. In other words, in these mutants budding will remain uninhibited (due to success of cell division) even when spindle is impaired. Three such
bub mutants allowed identification of three genes
BUB 1,
BUB 2 and
BUB 3 (budding uninhibited by benzimadazole in the mutants for these genes)
. Another set of five
'mad' mutants allowed identification of three more genes, namely
MAD 1,
MAD 2 and
MAD 3 (mitotic arrest deficient). Identification of so many genes suggest complex nature of this control, which allows proper spindle function, before mitosis is allowed to proceed.
What is the nature of this control which prevents mitosis without proper spindle function ? One possibility is that it operates by influencing inactivation of the p34
cdc2 kinase, normally required for successful completion of mitosis. If this inactivation of p34
cdc2 is not brought about, high p34
cdc2 activity will not allow the cell cycle to proceed from mitosis into interphase, until this activity drops. Whenever spindle is damaged either by chemicals or by mutation, p34
cdc2 activity rises, although in
bub and
mad mutants, the p34
cdc2 activity can not be kept up, so that the cells gradually exit from mitosis and enter interphase. The genes
BUB 2,
BUB 3 and
MAD 2 have been cloned and proteins encoded by them are being examined for their possible functions.
Gene for dependence of mitosis on growth
The product of
wee 1 gene is not essential for mitosis (in
S. pombe), but its presence delays mitosis. Deletion of
wee 1 or presence of its mutant allele gives cells smaller in size at mitosis and an increased dose of
wee 1 gives proportionately larger cells at mitosis. This suggests a genetic control of integration between growth and division.














