Yeast mating type switching
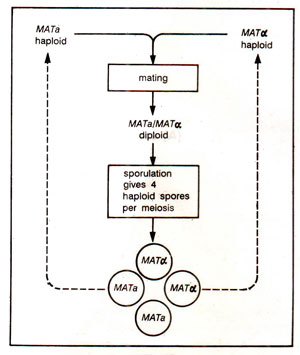
Life cycle of budding yeast, S. cerevisiae (Fig. 37.17) shows that there are two mating types (MATa and MATα), which fuse to give a diploid (MATa/MATα). The diploid cell sporulates and through meiosis forms four spores, two of each mating type. Each mating type is characterized by secretion of a pheromone (MATa secretes a factor and MATα secretes a factor) and a surface receptor for the pheromone of the opposite type. The diploids (MATa/MATα) produce no pheromone or surface receptor. The pheromone-receptor interaction activates a cell membrane protein called G protein consisting of three subunits (α β γ). The activation needs a GTP molecule, which displaces GDP associated with αβγ complex and releases αfrom βγ dimer.
This separation of a from βγ activates the next protein in signal transduction pathway. Therefore G protein is also called a transducin. The dimer βγ alone (and not α) is capable of bringing about the activation. The above information has come from a study of mutations (called STE = sterile), which are incapable of mating. Atleast ten such mutations (STE 1-7, 11, 12, 18) are known.
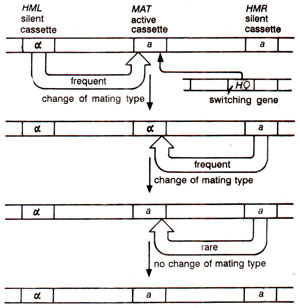 A remarkable feature in yeast is the ability of some strains (which carry dominant HO allele and not those which carry recessive ho allele) to switch their mating type from a to αand vice versa. Consequently an HO haploid strain, irrespective of its original mating type, will soon have cells of both mating types giving diploids (MATa/MATα). This is achieved by the presence of a MAT (a or α) locus with an active cassette (type a or α) associated with two silent cassettes at loci HML(α) on the left and HMR(a) on the right. Only the active cassette at MAT locus is expressed which can be MATa or MATα. This can be replaced by HMLαor HMRa respectively leading to change in mating type (a→α or α→a). MATa
is almost always replaced by, HML and MATα by HMR, so that if HML is mutated to HML a or HMR is mutated to HMRα, replacement by them does not change mating type (Fig. 37.18). The HML and HMR are constitutively repressed (rendered silent) by some repressors binding to silencer sequences, HML-E or EL and HMR-E or ER located ~ 1 kb upstream of these loci (as discussed earlier). The switching is achieved by the activity of an endonuclease coded by thedominant gene HO.
A remarkable feature in yeast is the ability of some strains (which carry dominant HO allele and not those which carry recessive ho allele) to switch their mating type from a to αand vice versa. Consequently an HO haploid strain, irrespective of its original mating type, will soon have cells of both mating types giving diploids (MATa/MATα). This is achieved by the presence of a MAT (a or α) locus with an active cassette (type a or α) associated with two silent cassettes at loci HML(α) on the left and HMR(a) on the right. Only the active cassette at MAT locus is expressed which can be MATa or MATα. This can be replaced by HMLαor HMRa respectively leading to change in mating type (a→α or α→a). MATa
is almost always replaced by, HML and MATα by HMR, so that if HML is mutated to HML a or HMR is mutated to HMRα, replacement by them does not change mating type (Fig. 37.18). The HML and HMR are constitutively repressed (rendered silent) by some repressors binding to silencer sequences, HML-E or EL and HMR-E or ER located ~ 1 kb upstream of these loci (as discussed earlier). The switching is achieved by the activity of an endonuclease coded by thedominant gene HO.

Fig. 37.18. Change of mating type in yeast due to replacement by active cassette by silent cassette of opposite type : when replacement involves cassettes of same type, the mating type remains unaltered.