Gene vs allele : A new concept of allelomorphism
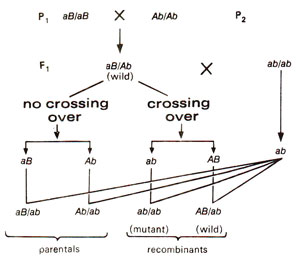
Fig. 14.1. Recombination test (a test cross) in which two linked genes in repulsion phase (aB/Ab)recombine and give rise to wild type individuals.
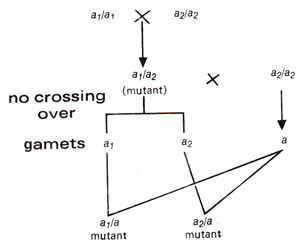
Fig. 14.2. A cross between two mutants of same gene showing lack of recombination as shown by absence of wild type individuals.

Fig. 14.3. A test of allelism (classical concept).
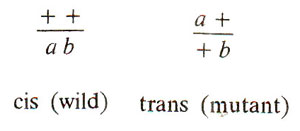
Fig. 14.4. Cis and trans arrangements of alleles a and b in heterozygous condition.
In classical genetics, a distinction was made between gene and allele on the basis of following two criteria.
Recombination test.
Recombination was believed to take place between two genes but not between two alleles. In other words, intragenic interallelic recombination was not conceived. For instance; a hybrid aB/Ab between mutants aa (aaBB)and bb (AAbb)for two linked genes A and B could give rise to wild type progeny on a test cross. Since A and B are linked, this would be possible only due to recombination (Fig. 14.1). On the other hand if two mutants a1a1 and a2a2 belonging to same gene A were crossed and F1 (a1/a2)is test crossed (a1a2 x aa)no wild type progeny would be expected (Fig. 14.2). It was thus, earlier believed that different genes or loci could recombine with each other by crossing over but different alleles of a gene could not. This test of allelism is illustrated in Figure 14.3.
It was also shown that mutant alleles of two different genes coming from two parents, thus being in repulsion phase (also known as trans configuration), will complement giving rise to wild type in F1 generation. But the mutant forms allelic to each other, will never complement (Fig. 14.4).

Fig. 14.1. Recombination test (a test cross) in which two linked genes in repulsion phase (aB/Ab)recombine and give rise to wild type individuals.

Fig. 14.2. A cross between two mutants of same gene showing lack of recombination as shown by absence of wild type individuals.

Fig. 14.3. A test of allelism (classical concept).
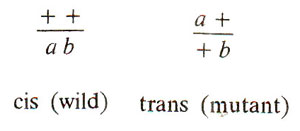
Fig. 14.4. Cis and trans arrangements of alleles a and b in heterozygous condition.




