Membrane Proteins
Approximately one third of all proteins are tightly associated with membranes. These are much more difficult to crystallize or study by NMR than water-soluble proteins. As a consequence, there are far fewer structures of membrane proteins. Even so, those that have been determined provide insight into the manner in which polypeptide chains interact with lipid bilayers.Membrane proteins fall into two classes: peripheral and integral. Peripheral membrane proteins are associated with the membrane, but may be removed by high concentrations of salt or metal chelators such as EDTA. In most aspects the structures of peripheral membrane proteins are very similar to water-soluble proteins. Integral membrane proteins differ in that they are very difficult to extract from the lipid bilayer and require detergents for solubilization. Detergents disrupt the lipid bilayer and bind to the hydrophobic surfaces of the protein that are buried within the membrane.
Integral membrane proteins all share the common problem of inserting a polypeptide chain into the hydrophobic interior of the lipid bilayer. This poses a thermodynamic problem on account of the hydrogen bonding propensity of the polypeptide chain. Clearly any segment of the protein that passes through the lipid bilayer must accommodate the hydrogen bonding potential of the polypeptide chain. Originally it was believed that an α-helix would be the only secondary structural element to pass through the lipid bilayer since it alone fulfills the hydrogen bonding capacity of the polypeptide chain in a consecutive manner. Indeed the α-helix is the only way to pass a single transmembrane segment of protein through a membrane. However, although a large number of membrane proteins are formed from α-helical bundles a significant number are built from β-strands. Both of these strategies for building integral membrane proteins present interesting biophysical problems.
A. α-Helical Membrane Proteins
Many fully inserted membrane proteins utilize bundles of α-helices tospanthe lipid bilayer. The structure of the photoreaction center, the first membrane protein whose structure was determined, clearly shows this strategy (Fig. 17a). It consists of polypeptide chains that span the lipid bilayer utilizing 11 transmembrane α-helices. Interestingly the surfaces that face the interior of the bilayer are more hydrophobic than the interior of the protein, whereas the components that face the aqueous environments are similar to the surfaces of water soluble proteins. Thus there is no tendency for proteins to unfold in the lipid bilayer. This suggests that the same forces that stabilize water-soluble proteins are responsible for the stability of membrane proteins. α-Helices are the major component of proton pumps such as bacteriorhodopsin (Fig. 17c) and K+ channel proteins.
B. β-Sheet Membrane Proteins
The outer membranes of gram-negative bacteria contain channels that allow the diffusion of small solutes and ions into the periplasmic space. These channels are formed by bacterial porins that are built almost entirely of antiparallel transmembrane β-strands (Fig. 17b). The topology of these proteins is exceedingly simple, consisting of upand- down strands where the first strand hydrogen bonds with the last strand of the sheet. In this way the hydrogen bonding potential of the β-strands is completely satisfied and a hydrophobic surface is presented by the side chains that extend into the lipid bilayer. The channel lies down the middle of the barrel. The side chains that line the pore provide some selectivity for the nature of the solutes that diffuse through the channel. All bacterial porins exist as oligomers (mostly trimers) where the interface between the porin subunits form a hydrophobic interior that is otherwise missing from these proteins.
This type of antiparallel packing of β-strands in integral membrane proteins has also been observed in hemolysin. Hemolysin is a heptameric pore forming protein from Staphylococcus aureus, where each of the subunits contributes two antiparallel β-strands to the transmembrane segment creating a 14 stranded barrel.
C. Other Membrane Motifs
Not all membrane proteins extend completely through the lipid bilayer, indeed many of the biosynthetic enzymes that are tightly bound to the membrane only extend into one face. A good example of this is prostaglandin synthase (Fig. 17d). This enzyme converts arachidonic acid into prostaglandin (PGH2) and exhibits two catalytic activities: a cycloxygenase and peroxidase. It is an important enzyme since it catalyzes a critical step in the biosynthesis of a wide range of eicosanoids that control the inflammatory response, pain and fever, blood pressure, induction of blood clotting, induction of labor, and the sleep/wake cycle. In most respects the structure of this dimeric enzyme is very similar to other water enzymes, except that it exhibits a highly hydrophobic surface that is built from four α-helices per subunit that are inserted into the lipid bilayer. The use of α-helices to penetrate one side of a lipid bilayer would appear to be a common feature of proteins that interact with one side of a membrane. Interestingly the active site opens to the lipid bilayer via a large tunnel. This provides a thermodynamically acceptable route for arachidonic acid, which is a hydrophobic substrate to enter the enzyme. Prostaglandin synthase is the site of action of nonsteroidal anti-inflammatory drugs such as aspirin and ibuprofen, which act to block the channel that opens to the lipid bilayer and prevent arachidonic acid from being converted to PGH2.
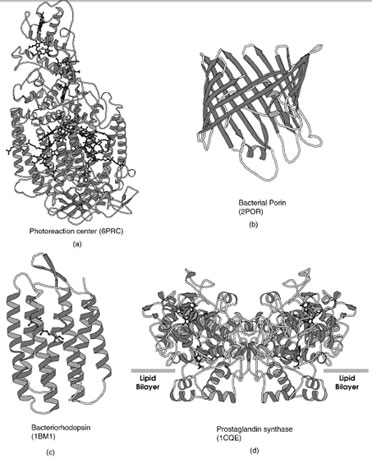 |
| FIGURE 17 Ribbon representation of selected integral membrane proteins. (a) Photoreaction center, (b) bacterial porin, (c) bacteriorhodopsin, (d) prostaglandin synthase. |




