Chromosome Inversions
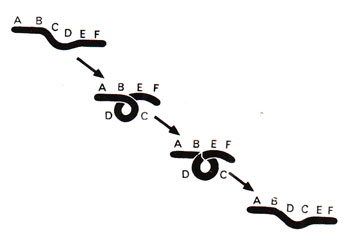
An inversion is produced when there are two breaks in a chromosome and the intercalary segment reunites in reverse order i.e. the segment rotates at 180°. Let us imagine that a chromosome 1-2-3-4-5-6-7-8 gives rise to another chromosome having the order 1-2-7-6-5-4-3-8. The segment 3-4-5-6-7 has rotated here at 180° giving an inverted order of genes 7-6-5-4-3. A similar hypothetical example using a chromosome ABCDEF has been shown in Figure 19.15, where due to coiling, breaks occur between B and C as well as between D and E. Reunion at broken ends may lead to inversion of the segment CD into DC.
The inversion can be of two types : (i) paracentric inversion, and (ii) pericentric inversion. Paracentric inversions are those inversions, where inverted segment does not include centromere. On the other hand, in a pericentric inversion, inverted segment includes centromere. In order to remember these terms and their meaning, one should bear in mind that pericentric means surrounding the centromere or on the periphery of centromere.
Cytology of inversions
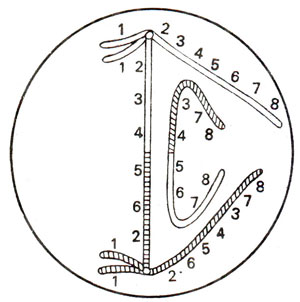
Due to an inverted segment in one of the two homologous chromosomes, the normal kind of pairing is not possible in an inversion heterozygote. In order to enable pairing of homologous segments, a shape of loop is formed by each of the two chromosomes as shown in Figures 19.16 and 19.20. This kind of configuration will be observed both in paracentric as well as in pericentric inversions. As will be observed, the products of crossing over and the subsequent stages of meiosis will differ in these two kinds of inversions.
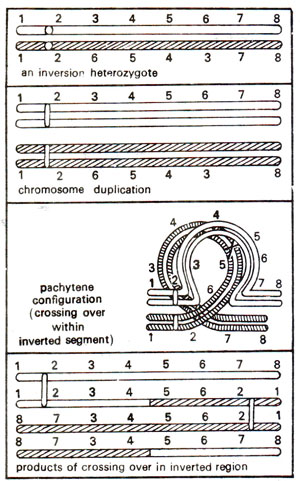
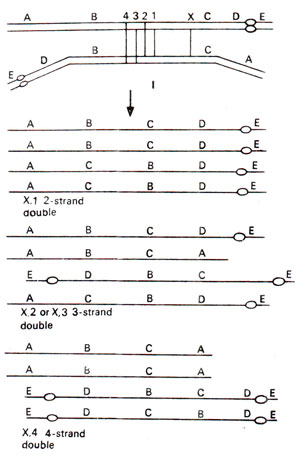
Paracentric inversion. A single crossing over or an odd number of crossovers in inverted region will result into formation of a dicentric chromosome (having two centromeres) and an acentric chromosome (with no centromere). Of the remaining two chromatids, one will be normal and the other will carry the inversion (Fig. 19.16). The dicentric chromatid and acentric chromatid will be observed at anaphase I in the form of a bridge and a fragment (Fig. 19.17). Double crossovers and crossovers within and outside inversion will give various kinds of deficiencies and duplications (Fig. 19.18). These will also give rise to a variety of characteristic configurations at anaphase I and anaphase II (Fig. 19.19).
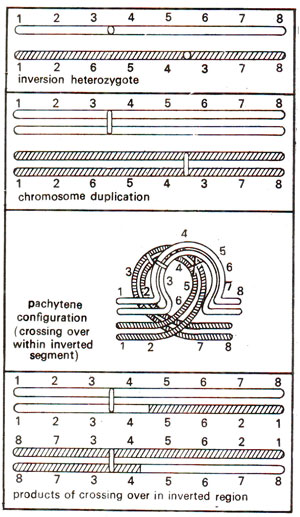
Pericentric inversion. In a pericentric inversion (where centromere is present within the inverted segment), the pachytene configuration observed is similar to the one described above for paracentric inversion (Fig. 19.16). However, the products of crossing over and configurations at subsequent stages of meiosis differ. In this case, two of the four chromatids resulting after meiosis will have deficiencies and duplications. However, unlike paracentric inversion, no dicentric bridge or acentric fragment will be observed (Fig. 19.20). Consequently, at anaphase I, no bridge or fragment will be seen.
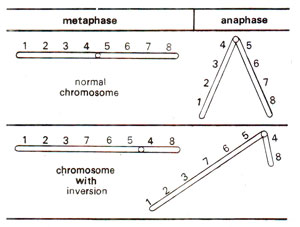
However, in pericentric inversion, if two breaks are not situated equidistant from the centromere, this will result in a change in shape of the chromosome. For instance, a metacentric chromosome (with centromere in the centre) may become submetacentric and vice versa (Fig. 19.21).

Genetic consequences of inversion
As discussed in the preceding section on cytology, among four chromatids resulting after crossing over, the two chromatids resulting from crossing over would have deficiencies and duplications. The gametes having these chromosomes will not function. Therefore, there should be considerable gametic or zygotic lethality. In plants, there will be sufficient pollen sterility. However, since the products of single crossover will not function and the only crossovers recovered will be double crossovers, the observed frequency of recombination between any two genes in question will be considerably reduced. Due to this reason, inversions, are often called crossover suppressors. This reduction in crossing over is not the actual reduction in cytological crossing over, but is the result of lack of recovery of the products of single crossovers. This property of inversions has been utilized in the production of ClB stock, used by H.J. Mulier for the detection of sex linked lethal mutations (Mutations : 1. Morphological Level (Including Lethal Mutations)).
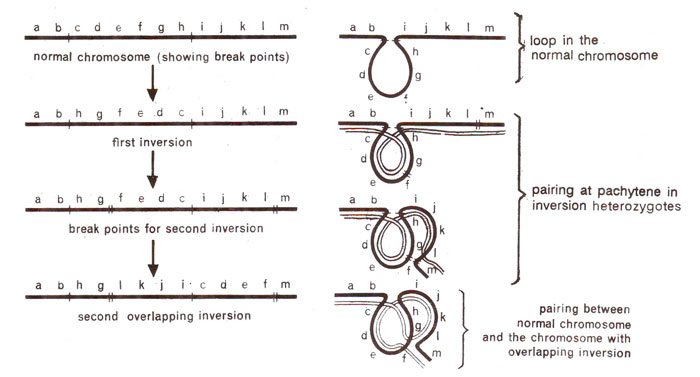
Overlapping inversions
Sometimes a second inversion is induced in a chromosome which already has one inversion. This results in an overlapping inversion, if the segments involved in first and second inversions contain a common region. The gene orders and meiotic configurations found in an inversion heterozygote of this type are shown in Figure 19.22.
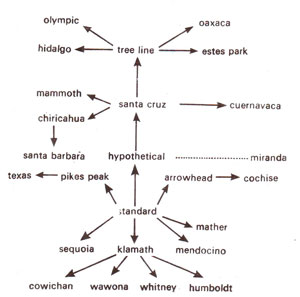
Inversions are known to have played a significant role in evolution of different species and races of Drosophila. This knowledge is particularly available in Drosophila due to the ease of identification of inversions in salivary gland chromosomes. Theyalso occur in plants, but can not be so easily worked out in the absence of giant chromosomes. In Drosophila, however, it is obvious that inversions occurred spontaneously in nature and became established in populations due to the adaptive benefit they conferred. Due to adaptive value, these inversions are restricted to definite localities and the different races actually derived their names after these localities, Th. Dobzhansky and A.H. Sturtevant even derived evolutionary relationships between races on the basis of overlapping inversions. These relationships are shown in the form of a tree in Figure 19.23.
Cytology of inversions
Due to an inverted segment in one of the two homologous chromosomes, the normal kind of pairing is not possible in an inversion heterozygote. In order to enable pairing of homologous segments, a shape of loop is formed by each of the two chromosomes as shown in Figures 19.16 and 19.20. This kind of configuration will be observed both in paracentric as well as in pericentric inversions. As will be observed, the products of crossing over and the subsequent stages of meiosis will differ in these two kinds of inversions.

Fig. 19.16. Chromosome pairing and products of crossing over in a paracentric inversion heterozygote.

Fig. 19.18. Consequences of 2-strands, 3-strands and 4-strands double crossovers within a paracentric inversion.

Fig. 19.19. On the top is shown inversion loop in an inversion heterozygote. Below are shown anaphase I (AI) and anaphase II (All) configurations (A-D) resulting from four different combinations of crossovers within and outside the inversion loop. A. single crossover at positions I or 2; B. 4-strand double crossovers at positions 1 and 2; C. 3-strand double crossovers, with one crossover at position 1 or 2 within inversion loop and the other crossover outside the loop at positions 3; D. 4-strand triple crossover at position 1, 2 and 3 (redrawn from McClintock, 1938).

Fig. 19.20. Chromosome pairing and the products of crossing over in a pericentric inversion heterozygote.
As discussed in the preceding section on cytology, among four chromatids resulting after crossing over, the two chromatids resulting from crossing over would have deficiencies and duplications. The gametes having these chromosomes will not function. Therefore, there should be considerable gametic or zygotic lethality. In plants, there will be sufficient pollen sterility. However, since the products of single crossover will not function and the only crossovers recovered will be double crossovers, the observed frequency of recombination between any two genes in question will be considerably reduced. Due to this reason, inversions, are often called crossover suppressors. This reduction in crossing over is not the actual reduction in cytological crossing over, but is the result of lack of recovery of the products of single crossovers. This property of inversions has been utilized in the production of ClB stock, used by H.J. Mulier for the detection of sex linked lethal mutations (Mutations : 1. Morphological Level (Including Lethal Mutations)).
Sometimes a second inversion is induced in a chromosome which already has one inversion. This results in an overlapping inversion, if the segments involved in first and second inversions contain a common region. The gene orders and meiotic configurations found in an inversion heterozygote of this type are shown in Figure 19.22.