Nucleic Acid Hybridization
From developments in the area of genetic engineering and molecular biology, a powerful tool known as DNA hybridization has emerged. This technique is used to detect the presence of DNA from pathogens in clinical specimens and to locate specific genes in cells. DNA hybridization takes advantage of the ability of nucleic acids to form stable, double-stranded molecules when two single strands with complementary bases are brought together under favorable conditions.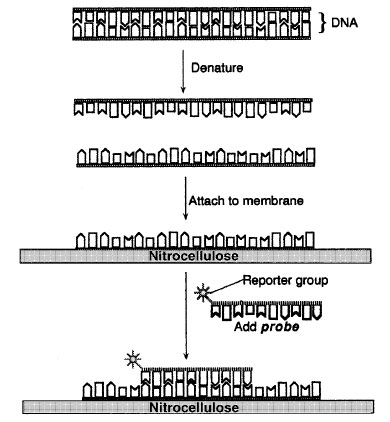 |
| Figure 9-1 DNA hybridization. |
In DNA hybridization assays, DNA from a virus or cell is denatured with alkali to separate the strands. The single strands of DNA are then attached to a solid support such as a nitrocellulose or nylon membrane so that the strands do not reanneal (Figure 9-1). The DNA is attached to the membrane by its sugar-phosphate backbone with the nitrogenous bases projecting outward. To characterize or identify the target DNA , a single-stranded DNA or RNAmolecule of known origin, called a probe, is added to the membrane in a buffered solution. This allows the formation of hydrogen bonds between complementary bases. The probe, so called because it is used to seek or probe for DNA sequences, is labeled with a reporter group, which may be a radioactive atom or an enzyme whose presence can be easily detected.
The probe is allowed to react with the target DNA ; then any unreacted probe is removed by washing in buffered solutions. After the washes, all that remains on the nitrocellulose is the target DNA and any probe molecules that have attached to complementary sequences in the target DNA , forming stable hybrids.
Hybridization of target and probe DNA s is detected by assaying for the probe's reporter group. If the reporter group is detected, hybridization has taken place. If no reporter group is detected, it can be assumed that the target molecule does not have sequences that are complementary to those of the probe, and hence, the gene or DNA segment sought is not present in the sample.
Three common formats are used in solid-phase hybridization assays; dot blot, Southern blotting, and in situ hybridization. In the dot blot assay, a specified volume of sample or specimen is spotted onto a small area of nitrocellulose membrane, which is then carried through the procedure described above. Southern hybridization assays (Figure 9-2) involve restriction enzyme digestion and agarose gel electrophoresis of the target DNA prior to the hybridization assay. The different bands on the agarose gel are transferred by capillary action onto a nitrocellulose or nylon membrane in a blotting apparatus. During the transfer, each of the DNA bands is transferred onto the membrane in the same relative position that it had in the gel. After the transfer, the target DNA is probed and detected, as in the dot blot assay. In situ hybridization assays involve the probing of intact cells or tissue sections affixed to a microscope slide. This type of solid- phase assay has the advantage that one cannot only detect the presence of target DNA in intact cells but also determine the location of such target DNA within a tissue. An important application of in situ hybridization is for the detection of viruses and certain types of bacteria within infected cells.
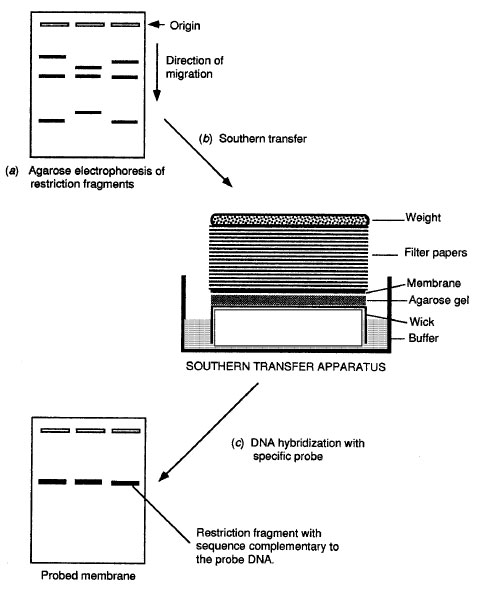 |
| Figure 9-2 Southern hybridization analysis. |
Notes
Four components of DNA hybridization:
1. Target DNA
2. Probe
3. Detection system
4. Format




