Multigene families with identical genes
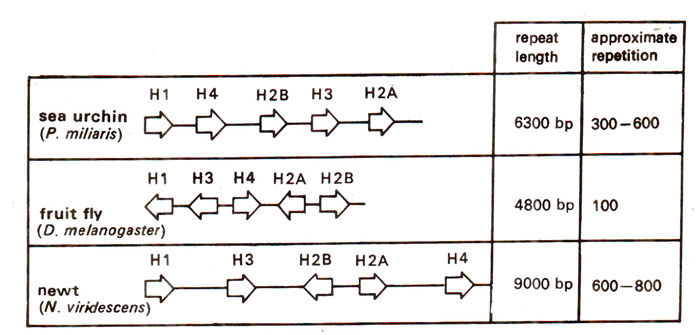
Fig. 44.5. Organization of highly repeated histone gene clusters in several organisms; genes are separated by non-transcribed spacer (NTS) regions; the direction of transcription is shown by arrows; the clusters are tandemly repeated in sea urchin and fruit fly, but not in the newt.
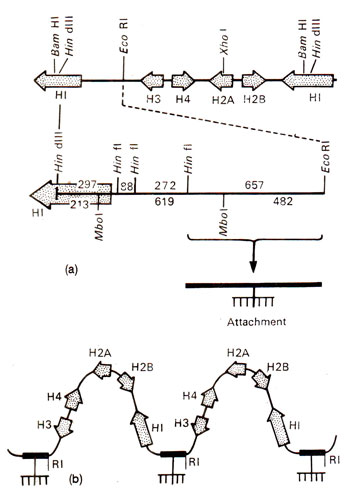
Fig. 44.6. (a) A restriction map of major histone gene cluster in Drosophila; each repeat unit has one specific attachment site to the nuclear scaffold, this attachment site is Hin I/Eco R I DNA segment, 657 bp long; (b) schematic representation showing how Drosophila histone repeats might be attached to the nuclear scaffold.
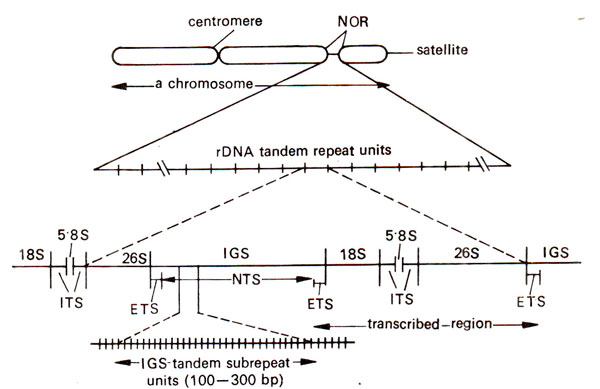
Fig. 44.7. Organization of ribosomal DNA (rDNA) in nucleolar organizing region (NOR) of a eukaryotic chromosome (IGS = intergenic spacer; NTS = non-transcribed spacer; ETS = external transcribed spacer; ITS = internal transcribed spacer; 18S, 5.8S, 26S = different genes responsible for 18S rRNA, 5.8S rRNA and 26S rRNA).
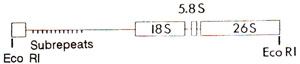
Fig. 44.8. An outline of the structure of rDNA repeat unit in wheat and barley.
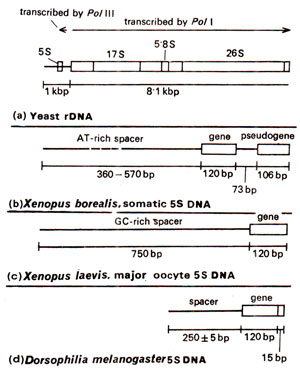
Fig. 44.9. Organization of 5S DNA (produces 5S RNA for ribosomes) in several organisms; only in yeast it is linked with repeat unit of rDNA shown in Figure 44.8; in other cases it is independent on a separate chromosome
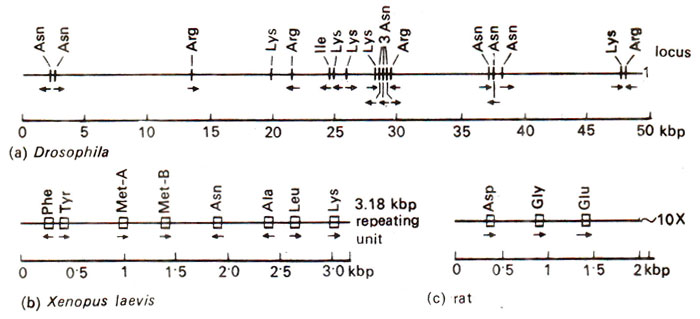
Fig. 44.10. Arrangement of tRNA genes in higher eukaryotes : (a) one of the 50-60 sites on Drosophila chromosomes where tRNA genes are clustered; (b)a 3.8 kilobase-pairs unit from Xenopus DNA, that is repeated about 150 times; (c) rat DNA showing only one of about 10 repeat units found in haploid genome.
Multiple copies of histone genes. The five major histone proteins, namely H1; H2A, H2B, H3, H4 are involved in the packing of DNA into nucleosomes, the chromatin subunits. When DNA is duplicated during S phase of the cell cycle, large quantities of these histone proteins are needed. To meet this demand, for each of the histone genes, there are present (i) 10-20 copies in birds and mammals and (ii) 600-800 copies in sea urchin and newt (amphibians). A higher number in amphibians suggests a response to their need for rapid cell division. The five genes for five histones form a basic unit that is repeated, although different genes within a repeat unit may differ in orientation (Fig. 44.5). These genes (coding sequences) in a repeat unit are highly conserved, but the spacer region differs in different organisms. The histone genes differ from most other eukaryotic genes in haying their transcripts devoid of introns and poly A tails.

Fig. 44.5. Organization of highly repeated histone gene clusters in several organisms; genes are separated by non-transcribed spacer (NTS) regions; the direction of transcription is shown by arrows; the clusters are tandemly repeated in sea urchin and fruit fly, but not in the newt.
Another interesting feature of the multigene family of histone genes observed in Drosophila is the presence of an 'attachment site' per repeat unit (Fig. 44.6), 100 such repeat units being present in each haploid genome. With the help of attachment site, the genes remain attached to nuclear scaffold (scaffold is the protein backbone of chromosomes, to which DNA remains attached). These attachment sites help in transcription.

Fig. 44.6. (a) A restriction map of major histone gene cluster in Drosophila; each repeat unit has one specific attachment site to the nuclear scaffold, this attachment site is Hin I/Eco R I DNA segment, 657 bp long; (b) schematic representation showing how Drosophila histone repeats might be attached to the nuclear scaffold.
Ribosomal RNA (rRNA) genes in tandem arrays. For the production of about 10 million ribosomes per cell in eukaryotes, rRNA genes are present in multiple copies at nucleolar organizing regions (NOR) of specific satellited chromosomes.
The number of these genes may vary from 50-30,000 in a cell and this number may be unequally distributed on NORs, if more than one such loci are present. The DNA comprising these genes is called rDNA (ribosomal DNA), which is repetitive in nature. Each repeat unit has (i) a coding region with genes that specify 18S, 5.8S and 28S rRNA molecules, (ii) a spacer region called intergenic spacer (IGS) and (iii) internal transcribed spacers (ITS) one each between 18S and 5.8S genes and between 5.8S and 28S genes. Since parts of IGS region adjoining the coding region, known as external transcribed spacers (ETS), is also transcribed, the use of the term non-transcribed spacer (NTS) for the whole spacer region has been considered to be inappropriate. Actually, the NTS makes only a part of the IGS (Fig. 44.7), the remaining part of IGS being ETS. The IGS in its turn has a region consisting of a tandem array of variable number of subrepeats ranging from 100-300 bp (bp = base pairs) in length. The variation in number and size of the subrepeats in IGS is responsible for variation in length of the repeat unit (IGS + coding region).

Fig. 44.7. Organization of ribosomal DNA (rDNA) in nucleolar organizing region (NOR) of a eukaryotic chromosome (IGS = intergenic spacer; NTS = non-transcribed spacer; ETS = external transcribed spacer; ITS = internal transcribed spacer; 18S, 5.8S, 26S = different genes responsible for 18S rRNA, 5.8S rRNA and 26S rRNA).

Fig. 44.9. Organization of 5S DNA (produces 5S RNA for ribosomes) in several organisms; only in yeast it is linked with repeat unit of rDNA shown in Figure 44.8; in other cases it is independent on a separate chromosome

Fig. 44.10. Arrangement of tRNA genes in higher eukaryotes : (a) one of the 50-60 sites on Drosophila chromosomes where tRNA genes are clustered; (b)a 3.8 kilobase-pairs unit from Xenopus DNA, that is repeated about 150 times; (c) rat DNA showing only one of about 10 repeat units found in haploid genome.
An unexpected feature of the DNA that is complementary to SnRNAs is that it consists of many more pseudogenes than the functional genes (10 : 1). For instance in humans there are 30 genes, for Ul (an SnRNA) on chromosome 1 and 500-1000 pseudogenes distributed throughout the genome.




