Bacterial Resistance to Antimicrobial Agents: Enzymatic
The activity of antimicrobial agents is usually very specific, affecting primarily essential bacterial cell structures or biochemical processes. For example, penicillin interferes with bacterial cell wall synthesis, gentamicin inhibits protein synthesis, and sulfonamides block folic acid synthesis. During the few decades of widespread antimicrobial agent usage, it has become evident that bacteria have the ability to inactivate or in some way circumvent the activity of almost every known agent. Resistance to antimicrobial agents can result from a mutation in a gene on the bacterial chromosome, or by acquisition from another organism of a plasmid (extrachromosomal DNA) that bears one or more “resistance” genes (R-factor). Acquisition of an R-factor can suddenly render a previously susceptible bacterium resistant to multiple antimicrobial agents. One of the most common mechanisms of bacterial resistance is the production of specific enzymes that destroy antimicrobial agents before they can affect the bacterium. For example, penicillinase is an enzyme that inactivates penicillin by breaking open a particular structure on the penicillin molecule called a beta-lactam ring (a synonym for penicillinase is beta-lactamase) (fig. 15.2). A gene on a plasmid in the bacterial cell provides instructions for formation of this enzyme. Although carriage of the penicillinase plasmid once appeared to be confined to certain strains of staphylococci and gram-negative bacilli, it is now found in some strains of bacteria that previously were considered to be universally susceptible to penicillin or its derivatives. These include Haemophilus influenzae, a cause of severe infections in children, before an effective vaccine became available, and Neisseria gonorrhoeae, the agent of gonorrhea.Bacterial enzymes can also be responsible for resistance to antimicrobial agents other than penicillin. Gentamicin and chloramphenicol, for example, may be inactivated by enzymes specific for these drugs, but there are additional mechanisms by which bacteria may resist the action of certain antimicrobial agents. These include alterations in critical bacterial enzymes or proteins such that they can no longer be directly affected by the drug; or changes in the bacterial cell wall or membrane that make the cell less permeable, preventing entrance of the agent.
Routinely, the clinical microbiology laboratory tests for bacterial susceptibility or resistance by the methods described in Experiments 15.1 and 15.2.
Alternatively, however, if you are interested only in the response of a given organism to a particular antimicrobial agent (e.g., Neisseria gonorrhoeae to penicillin), you can test the organism for its ability to
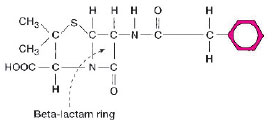 |
| Figure 15.2 Penicillin G (shown) and many of its derivatives are inactivated by a beta-lactamase (penicillinase). The enzyme breaks open the beta-lactam ring, which is a common part of the molecular structure of these antimicrobial agents. |
produce a sufficient amount of an enzyme that specifically inactivates tha drug. If the organism can be shown to possess the enzyme, it is considered to be resistant to the antimicrobial agent in question. One such test is illustrated in the following experiment, using a penicillinsusceptible organism and one that is resistant to penicillin because it produces penicillinase. The test uses a filter paper disk containing the chromogenic (color-producing) cephalosporin, nitrocefin. Like penicillin, the cephalosporins are degraded by betalactamases. When the test disk is inoculated with a penicillinase-producing organism, the yellow nitrocefin is broken down to a red end product.
| Purpose | To detect penicillinase production by a test bacterial strain |
| Materials | Filter paper disks impregnated with nitrocefin for performing the beta-lactamase test Sterile water or saline Clean glass slides Forceps Plate culture of a penicillin-resistant Staphylococcus aureus Plate culture of a penicillin-susceptible Bacillus subtilis |
Procedures
- Place two small drops of water or saline on the surface of a clean glass slide.
- Pick up a beta-lactamase disk with your forceps and place it in contact with one drop of fluid.
- Repeat this procedure with a second disk, placing it on the second drop of fluid. Do not oversaturate the disks.
- With your sterilized and cooled inoculating loop, pick up a portion of a B. subtilis colony and rub it across the surface of the first disk.
- Rub a portion of a S. aureus colony across the surface of the second disk.
- Observe the areas on the beta-lactamase disks where the organisms were inoculated for up to 30 minutes. A positive result is usually seen within 3 to 4 minutes.
Results
- A change in the color of the bacterial growth rubbed on the disk from yellow to red is a positive test indicating degradation of nitrocefin.
- Record your results in the chart.