Eukaryotic Gene Regulation
In eukaryotes, just as in bacteria, genes are regulated so that they are expressed at the right time and at the correct levels to maintain the cell or promote growth and proliferation. Cells in multicellular organisms must not only respond to the chemicals in their environment but must also work together through complex signaling that often affects gene activity.The controlling sites in eukaryotes are similar to those found in bacteria, however there are many more controlling sites and proteins affecting each eukaryotic gene. Transcription factors (TFs) attach to binding sites in promoter regions and stimulate RNA polymerase binding to the promoter site (Figure 4-3a,b). Transcription factors are produced constitutively since great numbers of genes depend on these for expression.
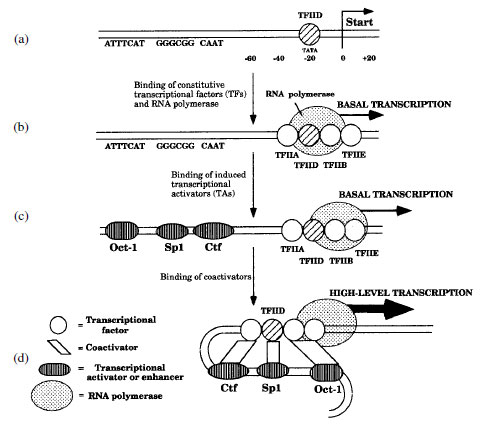 |
| Figure 4-3 Transcriptional activation in eukaryotes. |
Enhancers are bound by transcription activators (TAs) that are synthesized in response to specific signals. Most enhancers, such as the one that binds Gal4, are located hundreds or thousands of base pairs from the promoter sites. However, some induced activators, such as Fos and Jun, bind very near promoter sites. TAs cause the DNA to loop back on itself when they interact with the TFs near the promoter. This interaction between enhancer sites and initiator sites is usually necessary for transcription above a basal level (Figure 4-3c). Coactivators are activator proteins that often connect TFs and/or TAs with the RNA polymerase and may be essential for gene expression (Figure 4-3d).
Whereas only one RNApolymerase functions in bacterial cells, three different RNApolymerases are involved in eukaryotic nuclear transcription. The three polymerases initiate transcription only with specific combinations of TFs and TAs. RNA polymerase I transcribes the genes that specify 5.8S, 28S, and 18S rRNA. This polymerase is often found associated with chromosomes in the nucleoli. RNA polymerase II transcribes from promoters that control the synthesis of pre-mRNA, which consists of coding (exons) and noncoding (introns) regions. RNA polymerase III recognizes promoters that control the synthesis of relatively short RNAs such as tRNAs, 5S rRNA, and others.
In summary:
RNA pol I → rRNA
RNA pol II → mRNA
RNA pol III → tRNA (and 5S rRNA)
The binding of RNApolymerase to promoter sites is dependent on a number of TFs such as the TFIID complex (Figure 4-3a), which is functionally comparable to sigma factors in bacteria. TFIID is the first to bind close to the promoter at a site called a TATA box or Hogness box about -20 to -40 bp before the transcription start site. Once bound, TFIID helps organize other TFs required for initiation of RNA synthesis (Figure 4-3b). The complex of transcription factors and RNA polymerase comprise the preinitiation complex, which yields basal levels of transcription. Induction to higher levels requires the presence of other activators binding to enhancer elements. Activator proteins like Ctf, Sp1, and Oct-1 (Figure 4-3c) cause the DNA to loop back upon itself so that the activator proteins interact with the preinitiation complex, signalling the RNA polymerase to begin synthesizing high levels of RNA.




