Transcription Initiation and Termination
Transcription of a structural gene can only occur if there is a promoter for RNApolymerase binding near the beginning of the gene. In bacteria, promoters are usually 15-30 bp long. The base pair sequence of the promoter determines the efficiency with which RNA polymerase binds to it and thus the efficiency of transcription. In many cases, RNA polymerase binding to bacterial promoter sites is dependent upon a family of proteins called sigma (σ) factors. Each type of sigma factor determines the kind of promoter that RNApolymerase will recognize. Both a sigma factor and RNA polymerase are required for efficient binding to a promoter. After initiation, the sigma factor will dissociate from the RNApolymerase (see Figure 4-2a-c).Although prokaryotic promoters vary considerably, two short regions are shared in common by most promoters. Aregion on the nonsense strand about 10 bp before transcription initiates (-10 sequence) has a consensus 5'-TATAAT-3' and another region about 35 bp up (-35 sequence) has a consensus sequence 5'-TTGACA-3'. A consensus sequence contains the nucleotide sequence most commonly encountered in a wide variety of promoters.
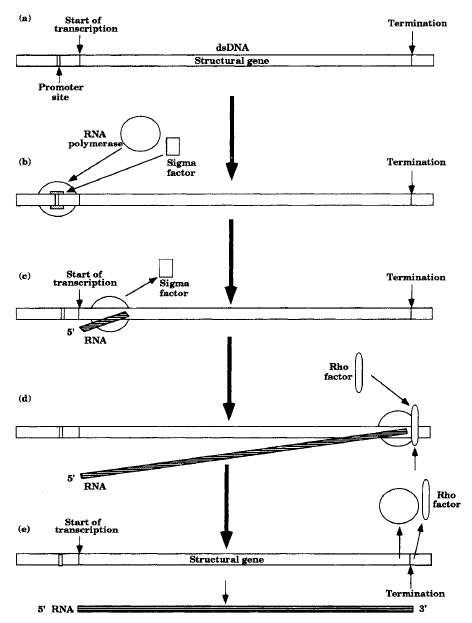 |
| Figure 4-2 Roles of Sigma and Rho. |
Most terminators at the end of a mRNA molecule code for a singlestranded region that folds on itself due to hydrogen bonding between complementary base pairs and codes for a final region containing many uracils. The hairpin structure that forms interacts with the RNA polymerase and stimulates its detachment from the DNA . Some terminators require terminator proteins such as Rho (ρ) to be active (Figure 4-2d,e). These Rho-dependent terminators lack a poly-U region in the 3' end of the terminator. The NusA protein is another termination factor that apparently binds directly to the RNApolymerase and catalyzes its release when it comes to termination sites.




