Evolution of a New Enzymatic Activity in Nature
Enzyme evolution in natural systems typically involves several steps: (1) gene
duplication, (2) change in functionality, and (3) selection for activity/specificity
(see Fig. 2.2). Duplications that occur at the individual gene level provide the
starting point for enzyme evolution.
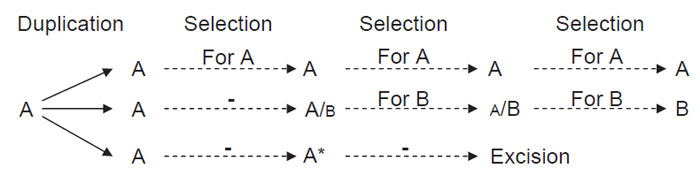 |
| FIGURE 2.2 General scheme for natural evolution of enzyme activity. A, Parental gene; A/B gene
encoding protein with dual activity that can perform activity B poorly; A/B, gene that encodes
protein with dual activity where B is the major activity; B gene encoding activity B that is unable to
perform activity A; A* represents a gene pseudogene that becomes excised. |
Mutations constantly arise in genes, but their accumulation depends on
stringency of the selection pressure for the function of the gene product. There
are three common fates that befall duplicated genes (Fig. 2.2): (1) retention of
function, (2) change of function (either change in activity or change in expression
pattern), or (3) loss of function followed eventually by excision.
Changes in enzyme function typically follow one of the three mechanisms
(Gerlt and Babbitt, 2001). The first mechanism is one in which a partial reaction
or a strategy for stabilization of energetically unfavorable transition state is maintained,
while the substrate specificity changes. In a second mechanism, substrate
specificity is maintained, but the chemistry changes during evolution. A third
mechanisminvolves retaining only the active site architecture, without maintaining
either substrate specificity or chemical mechanism.
Whichever of the mechanisms predominate, several features are likely to be
common. An initial gene duplication event is followed by the accumulation of
multiple mutations in one of the copies. A prerequisite for alteration of specificity
is that the original tight active site substrate specificity should relax allowing a
number of potential substrates to bind, or the same substrate to bind in alternate
conformations. Once an alternate substrate is capable of binding (or the same
substrate in a different binding conformation), an altered enzymatic transformation
may occur, resulting in the accumulation of a novel product. If the new
product conveys a selective advantage, over successive generations the accumulation
of further mutation/selection can lead to an increase in the new activity.
This ‘‘tuning’’ to the new substrate often occurs at the cost of catalytic efficiency
with respect to the original transformation. Thus, a characteristic of newly
evolved enzymes, or enzymes caught in transition, would be the observation of
relaxed specificity. Examples of this can be found in the fatty acid desaturases
(Broun
et al., 1998; Dyer
et al., 2002), where enzymes that exhibit ‘‘unusual’’
specificity with respect to the parental enzymes are often bifunctional in that they are capable of performing the archetypal reaction, often with lower catalytic
rates than the parental enzyme (Shanklin and Cahoon, 1998). Amino acid substitutions
that change the geometry of the binding pocket can be either direct, that
is, when the amino acid side chains directly line the binding pocket, or alternatively
can be at sites remote from the binding pocket and mediate their effects via
subtle changes in the relative organization of secondary structural elements.
In this context, amino acid side chains have been referred to as ‘‘molecular
shims’’(Whittle
et al., 2001) that orient the substrate with respect to the active
site in a very precise manner similar to the way carpentry shims are used to level
furniture. The stronger the selection pressure for the improvement in activity, the
faster it will progress.
Similarly, in the case of changes to the chemistry occurring on the same
substrate, it is envisaged that the enzyme became bifunctional with respect to
reaction outcome either by acquiring two or more alternate binding modes or by
alterations in the amino acid side chains that participate in catalysis. This has been
observed for the Fad2 family of fatty acid modification enzymes (Broadwater
et al.,
2002; Broun
et al., 1998). Once the new reaction occurs even at low levels, selection
can favor mutations that increase the new activity and lead to improved fitness at the organismal level and thus provide selective advantage. In either case where
substrate specificity changes, or chemistry on the same substrate alters, the ability
of an enzyme to perform alternate reactions shows it has the potential to acquire
a new dominant activity.
Duplicated genes that do not provide a selective advantage are rapidly excised
by unequal crossover at meiosis. Evidence for this includes studies in which
subfunctionalization is shown to occur rapidly upon polyploidization in cotton
(Adams
et al., 2003) and the observation of lower than expected occurrence of
pseudogenes (Force
et al., 1999).





