Recombination And/Or Introduction Ofsubsequent Mutations
Directed evolution experiments differ from traditional mutation-selection experiments
in that they typically involve cycles of improvement. This can be done in a
sequential fashion by identifying the most improved single variant and subjecting
it to further cycles of mutagenesis and screening/selection until a variant that
meets desired criteria is reached, or until further cycles fail to produce increases
in the desired property. However, this method tends to be slow and laborious.
A better method for recombining many improved variants is known as gene
shuffling (Stemmer, 1994b). This method is a variation on the mutagenesis method
described above in which genes are partially digested with the use of DNase and
subsequently reassembled by primerless PCR, except that a pool of improved
variants are used for the starting material rather than a single gene. The result
of this procedure is to make a new library of variants in which mutations
from different improved variants are recombined in many permutations and
combinations. In some variants, different positive amino acid changes that independently
improved the property of interest provide either additive or multiplicative
improvements in performance. In other cases, positive mutations could be partially obscured by negative mutations, so the process of improvement involves
both summation of positive mutations and, at the same time, elimination of
negative mutations. The removal of negative mutations can also be achieved
by backcross PCR. This technique is analogous to a traditional genetic backcross
experiment, but in this case, the improved variant is recombined with a molar
excess of parental gene, and the resulting variants screened for activity.
By
performing several cycles of this procedure, typically 4–7 rounds of screening/
recombination of improved variants, improvements in performance of 101–104
have been documented. Examples of successes using this technique include
conversion of a galactosidase into a fucosidase (Zhang
et al., 1997), increasing
the activity of a thermophylic enzyme at low temperatures (Merz
et al., 2000), and
the evolution of antibody-phage libraries (Crameri
et al., 1996).
While single gene shuffling is capable of generating huge changes in activity
with regard to specific substrates, it is still a fairly inefficient process. The reason
for this is that the variability introduced into the initial gene is somewhat limited
by the particular method of mutagenesis employed. A major advance in efficiency
of gene shuffling was made by incorporating several genes rather than a single
gene as a starting point, a process referred to as family shuffling (Crameri
et al.,
1998). In this method, several homologues (e.g., α, β, and γ of Fig. 2.3) with >50%
sequence identity are shuffled together and the resulting variants screened
for activity as described for single gene shuffling. An extension of this method
is called synthetic shuffling in which information from sequence comparisons
is used to generate oligonucleotides so that every amino acid from a set of
parents is allowed to recombine independently (Ness
et al., 2002).
Synthetic
shuffling has the advantage that shuffling can be used to exploit sequence information and does not require all of the physical genes to be in hand to perform
the experiment. Improvements employing these methods are shown to be more
rapid and larger in magnitude. The rational for this is that each homologue
represents a variant on the same protein fold and that during natural evolution
each of the genes has accumulated different positive sequence attributes that
contribute to the overall enzyme performance. During family shuffling, there is
the potential to sum these positive attributes to produce rapid increases in performance.
Another way to think of this is that a single gene shuffling experiment is
essentially starting off at a single point and radiating from there in sequence
space. For multigene shuffling, one starts with several independent points in
sequence, space, and combinations of each of the genes cover a larger portion of
sequence space than could be achieved from a single point (Fig. 2.3A).
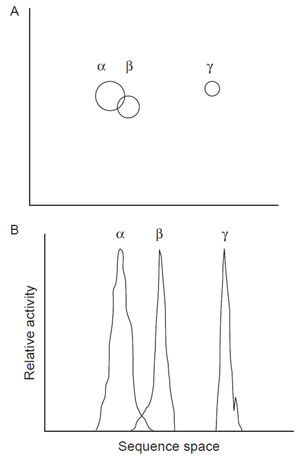 |
| FIGURE 2.3 (A) Sequence space; (B) Fitness
landscape. α, β, and γ represent enzymes with
different activities. |
Subsequent rounds of recombination and screening occur as before for single gene shuffling.
An important and intriguing finding from family gene shuffling experiments is
that when genes are shuffled, instead of getting activities that are intermediate
between the members shuffled, new activities beyond the range of the individuals
are identified. This is true for not only activities but also for qualitative parameters
such as range of region specificities. This has far-reaching implications in that new
diversity of biocatalysts can actually arise for parameters previously not found in
nature. The necessary criteria for exploiting this phenomenon are to identify and recombine the optimal parental genes and to have in place a robust high throughput
screen that has an excellent signal-to-noise ratio for the property of
interest. In addition to DNaseI-based recombination techniques, there are other
effective methods such as staggered extension process StEP PCR (Aguinaldo and
Arnold, 2003).
An interesting variation on single and multiple gene shuffling is that of
pathway and whole organism shuffling (Crameri
et al., 1997; Zhang
et al., 2002).
These broader-scale methods allow changes in regulatory elements, in addition to
changes in the coding regions to contribute to improved activity.





