Enzymes
Exonucleases
These enzymes act upon genome and digest the base pairs on 5' or 3' ends of a single stranded DNA or at single strand nicks or gaps in double stranded DNA (Fig. 3.1A).
Endonucleases
They act upon genetic material and cleave the double stranded DNA at any point except the ends, but their action involves only one strand of the duplex (Fig. 3.1B).
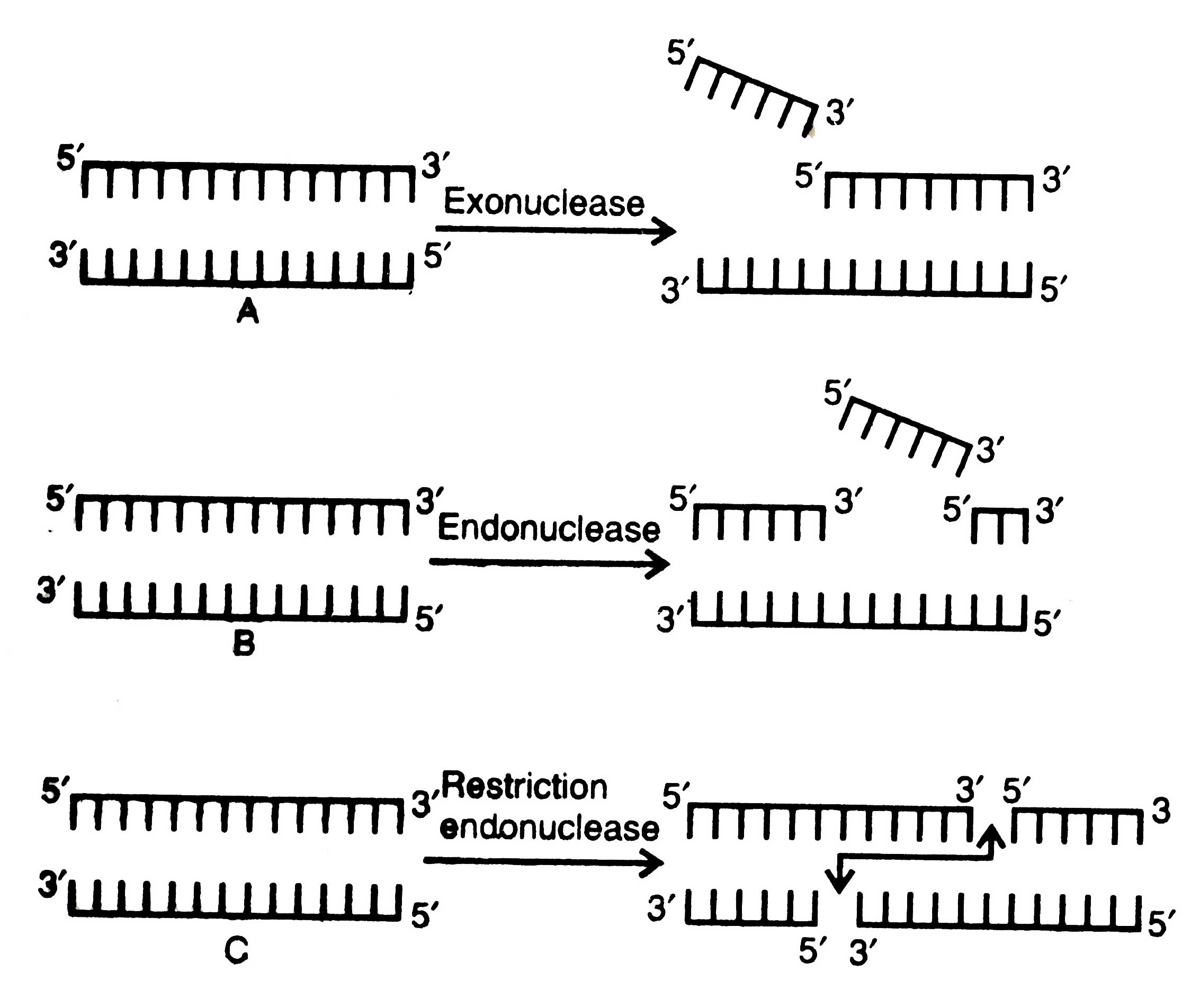
Fig. 3.1. Actionof DNA cleaving enzymes; A – exonuclease; B – endonuclease; C – restriction endonuclease.
These enzymes occur naturally in bacteria as a chemical weapon against the invading viruses and cut both strands of DNA when certain foreign nucleotides are introduced in the cell. These enzymes cleave a DNA to generate a nick with a 5' phosphoryl and 3' hydroxyl termini. There are two major restriction enzymes: Type I and Type II. Type f enzymes recognize a specific sequence of DNA molecule but cut elsewhere, and Type II make cuts only within the restriction sites and produce two single strand breaks, one break in each strand. Type II enzymes are the most important ones. As a result of their action the broken nucleotides form a DNA duplex which exhibit two fold symmetry around a given point. In some cases, cleavage in two strands are staggered to produce single stranded short projections opposite to each other with blunt or mutually cohesive sticky ends which are identical and complementary to each other (Fig. 3.1C).
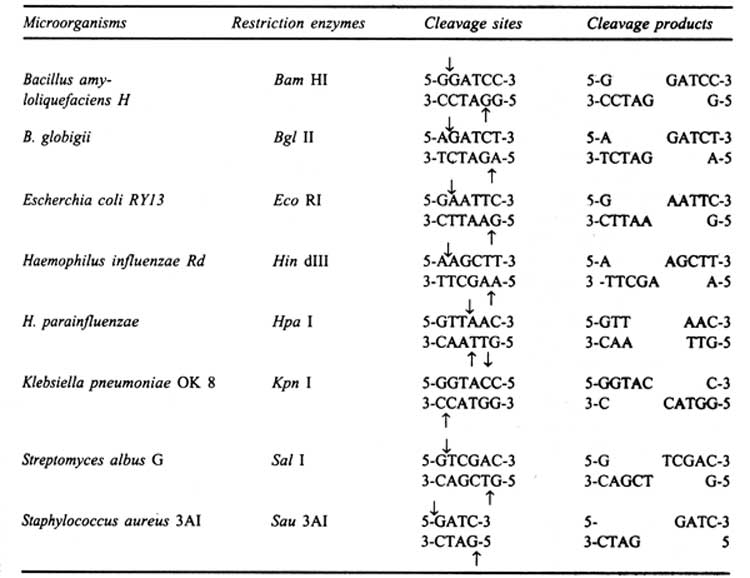
Roberts (1983) has given an extensive list of restriction enzymes and the sequences recognized by them. Most of the enzymes share a common central nucleotide in their recognition sequence for example, BamHI, BglII, Sau3A. But these enzymes recognize different sites in DNA and produce identical single stranded 5' tails which allow the joining of fragments generated by different enzymes within this set. The identical nature of the termini of cleaved DNA fragments from any organism is the very property which permits the annealing and subsequent ligation of the DNA from diverse sources (Glover, 1984).
Table 3.1. List of suppliers of restriction enzymes.
| Name of Companies |
|
| 1. | Benthesda Research Laboratories 411 North Stoncstrcet Avenue PO BOX 6010, Rockvile Maryland, 20850, U.S.A. |
| 2. | P.L. Biochemical Inc. 1037 West McKinlcy Avenue Milwaukee, Wisconiin, 53205, U.S.A. |
| 3. | Collaborative Research Inc.1365, Main Street,Waltham Massachusetts, 02154, U.S.A. |
| 4. | Boehringe Mannheim Gmbh Biochema 6800 Mannheim 31, West Germany (now Germany) |
| 5. | Miles Research Products PO Box 37, Stoke Poges Slough, Berkshire SL2 4LY, U.K. |
| 6. | Bangalore Genei Pvt. Ltd8th Main Road, Malleswaram Bangalore-560 055, India. |
The restriction enzymes are named based on some of the following principles:
(i) Name of the organism is identified by the first letter of the genus name and the first two letters of the species name to form a three letter abbreviation in the italic, for example, E. coli = Eco and H. influenzae = Hin, etc.
(ii) A strain or type identified is written as subscript e.g. EcoK for E. coli strainK, Hind for H. influenzae strain Rd.
(iii) In such cases where the restriction and modification systems are genetically specified by a virus or plasmid, the extra chromosomal element is identified by a subscript e.g. Eco RI, EcoPI, etc.
(iv) When a strain has several restriction and modification systems, these are identified by Roman numerals, for example Hindi, Hindll, Hindlll for H. influenzae strain Rd. etc. These Roman numerals should not be confused with those in the classification of restriction enzymes into Type I.