Cloning Vectors
Plasmids
Plasmids are the extrachromosomal, self-replicating, and double stranded closed and circular DNA molecules present in the bacterial cell. Plasmids contain sufficient genetic informations for their own replication. A number of host properties are specified by plasmids, such as antibiotic and heavy metal resistance, nitrogen fixation, pollutant degradation, bacteriocin and toxin piroductioivcolicin factors and phages (Dahl et al, 1981). Naturally occurring plasmids can be modified by in vitro techniques. Cohen et al. (1973) for the first time reported the cloning DNA by using plasmid as vector.
- It can be really isolated from the cells,
- It possesses a single restriction site for one or more restriction enzyme(s),
- Insertion of a linear molecule at one of these sites does not alter its replication properties,
- It can be reintroduced into a bacterial cell and cells carrying the plasmid with or without the insert can be selected or identified (Bernard and Helinski, 1980),
- They do not occur free in nature but are found in bacterial cells.
In many cases the principal objective of cloning experiment is the insertion of a particular restriction fragment into a suitable plasmid vector and its amplification. Amplification is a process of increasing the number of plasmid in a bacterial cell. In this process, a cell containing a relaxed plasmid is treated with a drug to inhibit protein synthesis.
| Cloning vectors | Natural occurrence | Size (Kb)* | Selective marker** |
| Plasmids | |||
| pACYC 177 | Escherichia coli | 3.7 | Ampr, Kanr |
| pBR 322 | E. coli | 4.0 | Ampr, tctr |
| pBR 324 | E. coli | ,8.3 | Ampr, tetr, El imm. |
| pMB9 | R. coli | 5.8 | Tcf |
| pRK 646 | E. coli | 3.4 | Ampr |
| pC194 | Staphylococcus aureus | 3.6 | Eryr |
| pSA 0501 | S. aureus | 4.2 | Strr |
| pBS 161-1 | Bacillus subtilis | 3.65 | Tetr |
| pWWO | Pseudomonas putida | 117 | Kanr |
| Cosmids | |||
| pJC 74 | Derived plasmid from Col EL | 16 | Amp1, El imm. |
| pJC 720 | do | 24 | El umn./Rifr |
| pHC 79 | Derivative of pBR 322 | 6 | Ampr, Tetr |
| Viruses | |||
| SV40 | Mammalian cells | 5.2 | - |
| Phage M13+ | E. coli | 6.4 | - |
| Phage l | E. coli | 4.9 | - |
Kanamycin (Kanr), rifampicin (Rifr), and colicin production (EL imm.) A physical map of plasmid pBR 322 is shown in ( Fig. 3.4.) The pBR 322 is constructed from the plasmids of E. coli, pBR318 and pBR320. It contains origin of replication (ori) that was derived from a plasmid related to naturally occur ring plasmid Col El. Therefore, its replication may be more faster than bacterial DNA. It also possesses genes conferring resistance to antibiotics e.g. ampicillin (amp1) and tetracycline (tef), and unique recognition sites for 20 restriction endonucleases. Certain restriction sites for example, Bam HI in the tetr genes of the plasmid are present within the gene in such a way that the insertion of foreign segment of DNA will inactivate the tef gene. The recombinant plasmid will allow the cells to grow only in the presence of ampicillin but will not protect them against tetra-cycline. The presence of an antibiotic resistant gene in a plasmid of bacterium will confer resistance to that antibiotic.
Therefore, the antibiotic resistant cells can be selected by culturing the cells on nutrient medium supplemented with ampicillin or tetracycline.
Species of Pseudomonas consist of a variety of plasmids which encode enzymes capable of degrading an enormous range of natural and synthetic organic compounds. The most studied plasmid among them is pwwo from P. putida. This is one member of a set of plasmids which have required the generic name TOL plasmids. It carries the genes that utilize toluene, m-and p-xylene, 3-ethyltoluene and 2,4- trimethyl benzene as well as their alcohol, aldehyde and carboxylic acid derivatives (Glover, 1984).
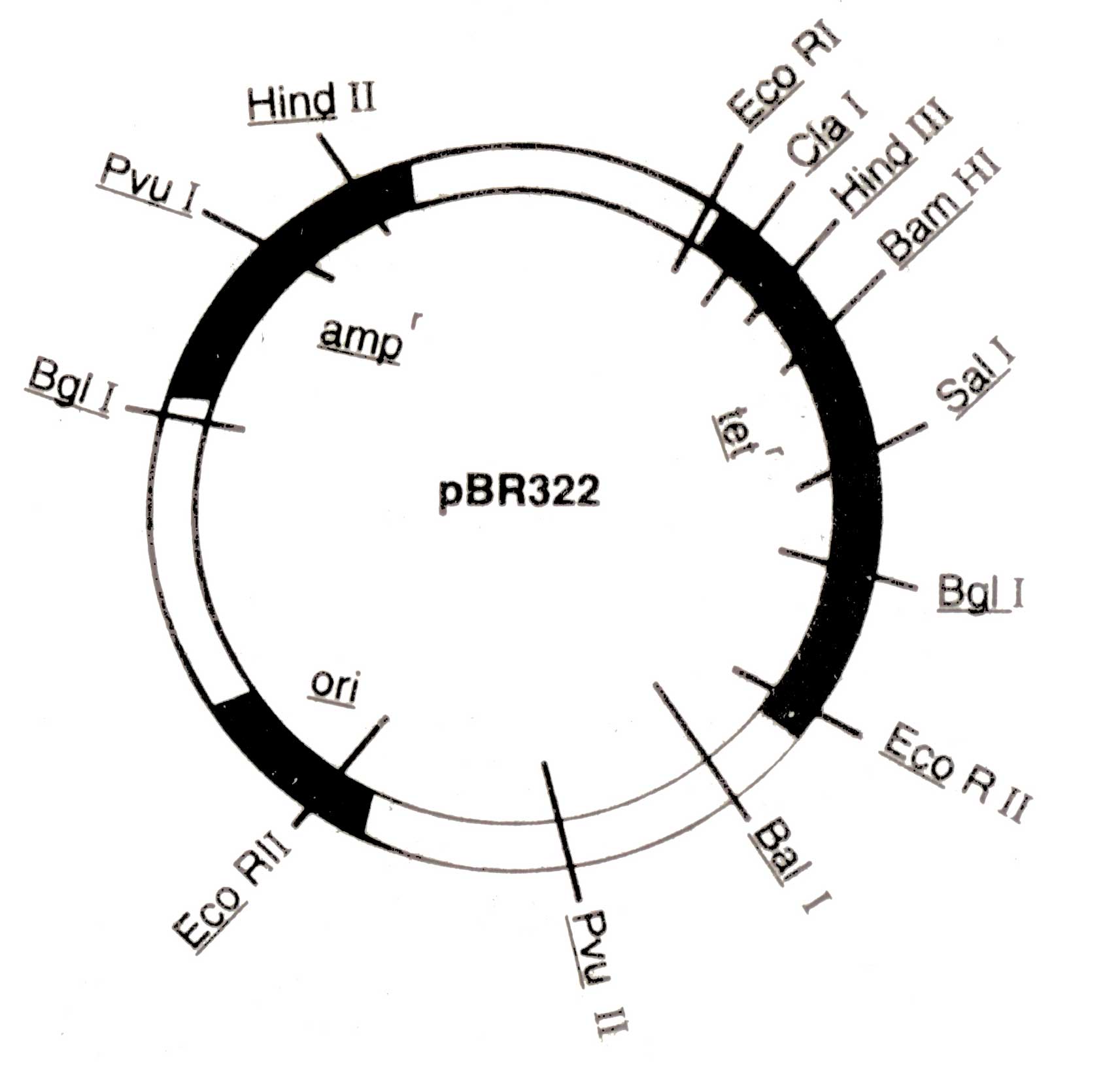
Fig 3.4. A map of pBR322 (4Kb) showing a number of restriction sites and regions encoding for resistance to ampicillin (ampr) and tetracycline (tetr), and origin of replication (ori)
Prof. Anand Mohan Chakrabarty and coworkers (1979) at the University of Illinois, USA. have developed strains of Pseudomonas which can be applied for degradation of synthetic organic compounds and pesticides. They are also making efforts to isolate bacterial strains that could reduce the viscosity of heavy oils by degrading the constituents of oils, waxes and paraffins. Thus, plasmids are an excellent vector for small fragment of DNA and, therefore, a necessary tool for establishing the cDNA bank.
Isolation of bacterial plasmid
Following are the steps for isolation of bacterial plasmids:
- Treat the prokaryotic cells with detergent. Cell membrane is solubilized. Plasmid DNA, chromosomal DNA and the other molecules are released. This mixture is known as lysate.
- Treat the lysate with potassium acetate/acetic acid solution. Chromosomal DNA with some protein molecule is precipitated.
- Remove the precipitate from lysate by centrifugation at high speed. Clear lysate is left. It contains plasmid DNA along with contaminating RNA, protein and a small amount of chromosomal DNA debris.
- Treat the lysate with-RNAase. Consequently, RNA is digested,
- Mix the lysate with phenol. Phenol and water is separated into layers. Phenol layer contains the contaminating proteins and RNAase, and water based lysate layer contains plasmid DNA.
- Remove the phenol layer. Precipitate the water based layer with alcohol as the genomic DNA is removed. Make use of it as desired.
Bacteriophages
A bacteriophage is virus which eats upon bacteria. A number of such viruses of different genetic material have been reported, for example ΦX174, phage λ, MI3, Fd11, R209, etc. Unlike plasmid vectors, the phage vectors are required for cloning of large DNA fragment and, therefore, the gene bank or genomic libraries can be constructed. They are an alternative to bacterial plasmids, and related cosmids. The process of infection and replication of phage in E.coli cell are shown in (Fig. 3.5).
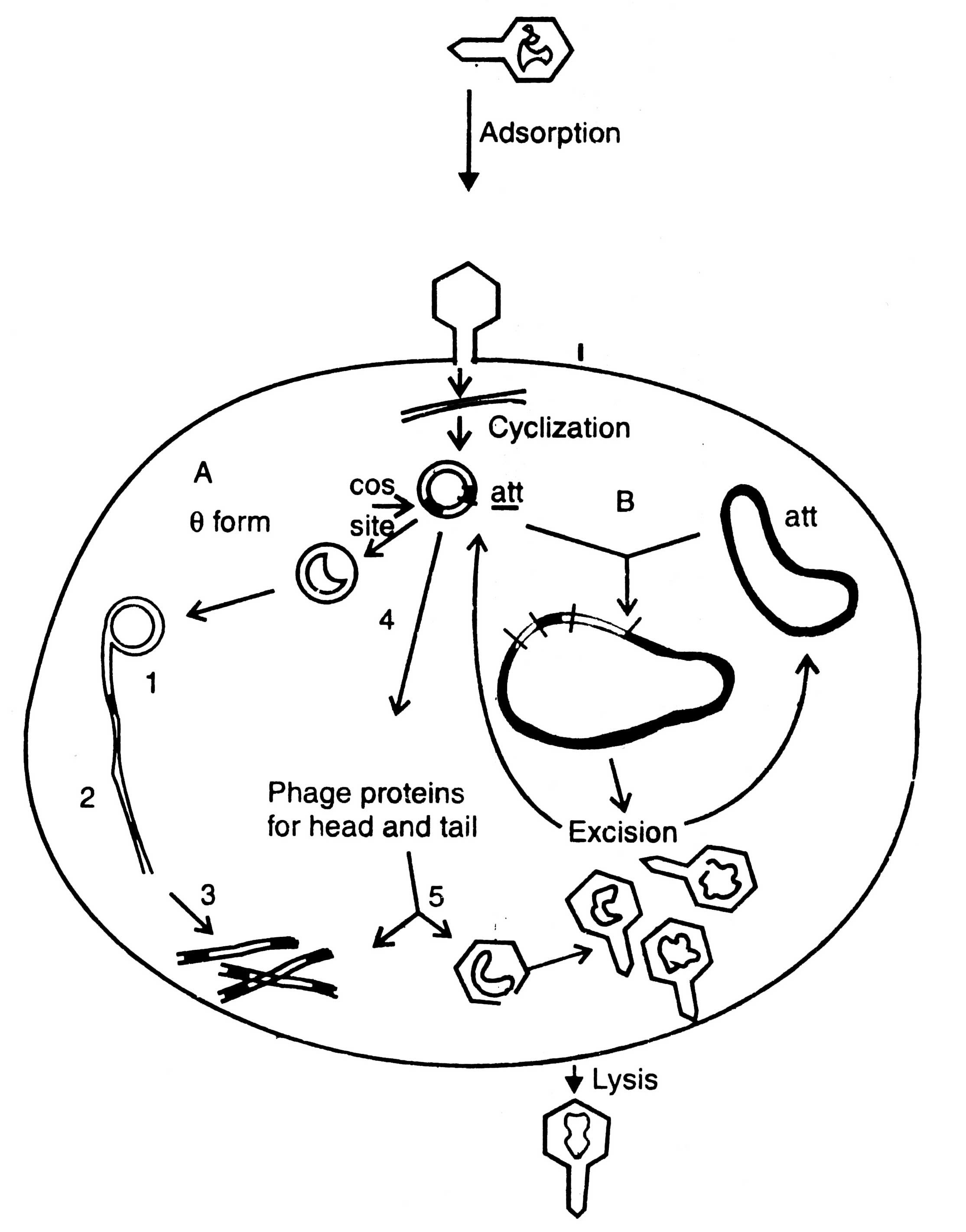
Fig. 3.5. Replication of phage lambda in a Escherichia coli cell. A - lytic cycle; 1. rolling circle replication; 2. production of concatemers; 3. cleavage at cos site; 4. transcription and translation; 5. packaging. B - lysogenic cycle.
Non-essential DNA sequence, therefore, is replaced by donor DNA and the inserted DNA propagated as phage lrecombinant with no detrimental to its replication or packaging.
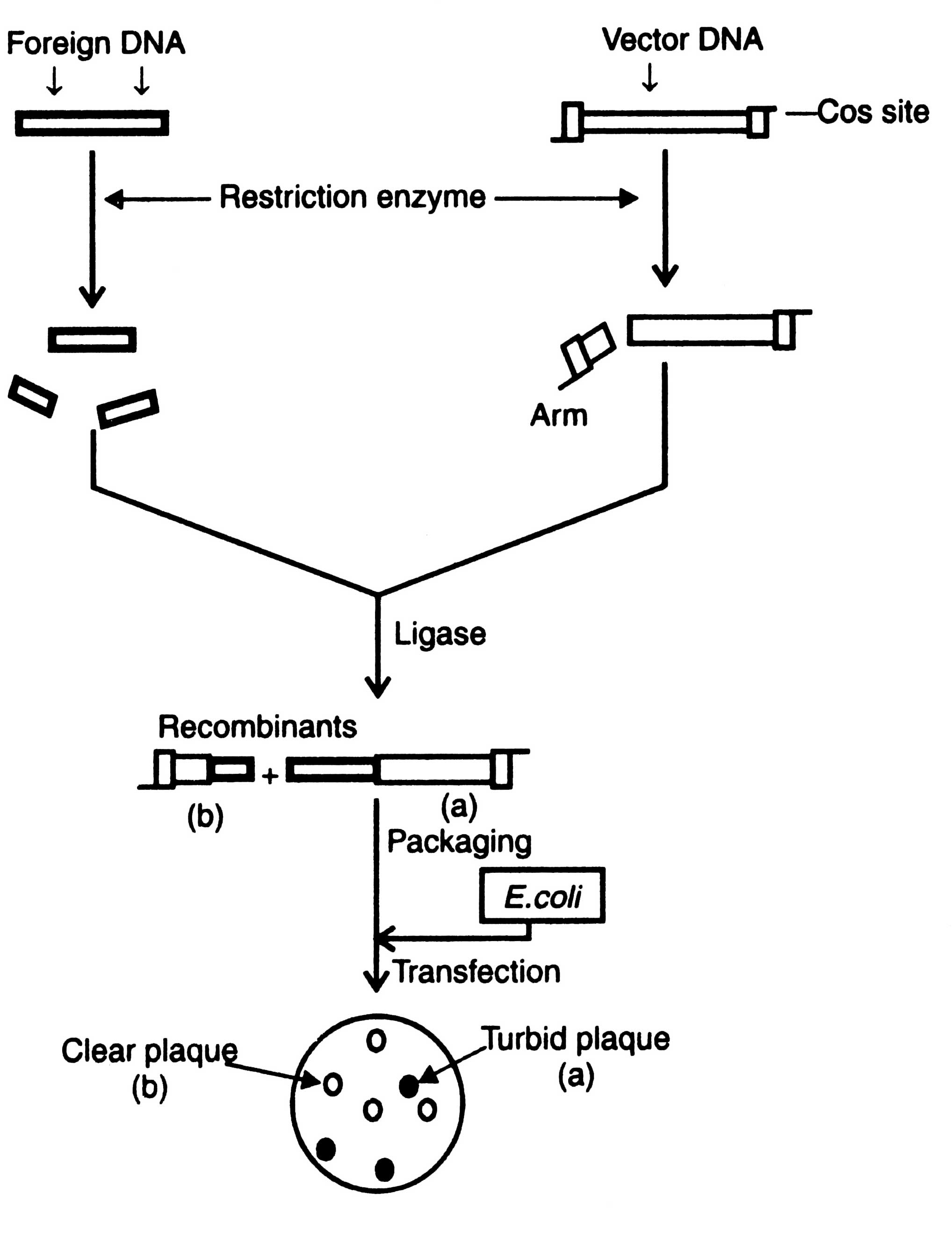
Following are the advantages of phage cloning system over the plasmids: (i) DNA can be packed in vitro into phage particles and transduced into E. coli with high efficiency, (ii) foreign DNA upto 25 Kb in length can be inserted into phage vector, and (iii) screening and storage of recombinant DNA is easier (Dahl et al, 1981). Before using the phage λ as vector, it is essential to remove the genome from the restriction sites for the enzymes commonly used for cloning. The restriction sites are eliminated by mutation or deletion before obtaining an useful cloning vector. Two types of phage cloning vectors have been constructed : the insertion vector and the replacement vector. Cloning procedures of both the vectors are shown in Pigs. 3.6 and 3.7.
Insertion vector
Insertion vectors have unique cleavage site into which relatively small piece of foreign DNA is inserted. Foreign DNA fragment does not affect the function of phage. The upper and lower limits of size of DNA that may be packed into phage particles is between 35-53 Kb. Therefore, the minimum size of vector must be above 35 Kb. It means the maximum size of a foreign DNA to be inserted is about 18 Kb. Cloning of an insertion vector is shown in Fig. 3.6.
Replacement vector
The replacement vectors have cleavage sites present on either side of a length of non-essential DNA of phage. As a result of cleavage left and right arms are formed, each arm has a terminal cos site and longer a stuffer region, the non-essential region, which can be substituted by foreign DNA fragment Fig. 3.7.
The maximum size of inserted DNA fragment depends on how much of the phage DNA is non-essential. It has been found that about 25-30 Kb of genome codes for essential products for lytic cycle. The remaining 20-25 Kb of genome could be replaced with the foreign DNA fragments of known essential products. The substituted vectors are gt, WES and λ 1059 (Dahl et al., 1981).
Non-essential part of the λ genome can be separated from the arms by electrophoresis or velocity gradient ultracentrifugation that facilitates to make size differences. Formation of multiple inserts can be checked by using alkaline phosphatase before ligation with insert fragment. Recombinant DNA formed by multiple inserts has too large genome to be packed in viral head. However, after ligation the recombinant DNA molecules have left arm plus large insert plus right arm linked by their cos sites at the arms.
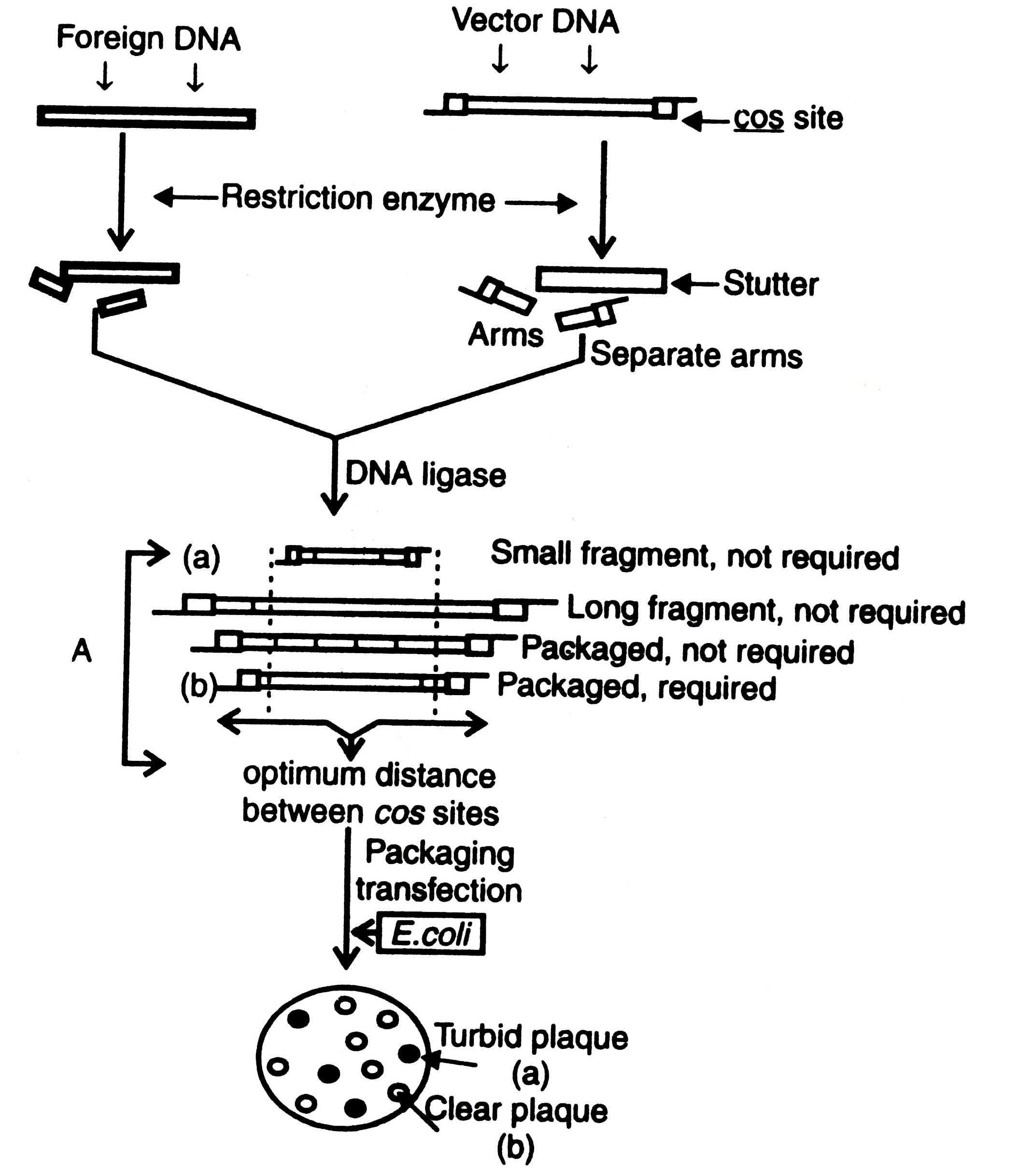
Fig. 3.7 Cloning of a replacement vector. A - optimization of the suitable sized ligation products for efficient packaging in phage.
Optimum distance from cos sites governs efficiency for packaging in E. coli. As a result of ligation the size of recombinant DNA fragment may be less or more than the required size or may have more than two cos sites of many small fragments of foreign DNA. Those having the range of viral head can be packed in vitro using a preparation of head and tail proteins. The viruses thus constructed are allowed to multiply in E. coli. Development of plaque turbidity is a useful criteria for the selection of recombinant phages. Plaque turbidity is determined by the presence of nonlysed bacteria. The recombinant phages give clear plaques due to inactivation of cl gene. By using these methods the clones containing recombinant DNA can be isolated from the wild type clones i.e. turbid plaques. The constructed genome has all the informations required for DNA replication and synthesis of the viral protein. The inserts can be identified by colony hybridization technique as described in Techniques of Genetic Engineering.
Cosmids
Based on the properties of DNA and Col El plasmid DNA, a group of Japanese workers (Fukumaki et al., 1976) showed that the presence of a small segment of phage lDNA containing cohesive end on the plasmid molecule is a sufficient prerequisite for in vitro packaging of this DNA into infectious particles. The cosmids can be defined as the hybrid vectors derived from plasmids which contain cos site of phage λ. For the first time it was developed by Collins and Hohn (1978).
Cosmids lack genes encoding viral proteins; therefore, neither viral particles are formed with the host cell nor cell lysis occurs. Special features of cosmids similar to plasmids are the presence of (i) Wigin of replication, (ii) a marker gene coding for antibiotic resistance, (iii) a special cleavage site for the insertion of foreign DNA, and (iv) the small size.
The cosmids have a length of about 5 Kb, the upper size limit of the foreign DNA fragment that may be inserted in cosmids and packed into phage particles is, therefore, approximately 45 Kb, much larger than it would be possible to clone in phage X or plasmid vector (Dahl et al., 1981). According to the size of cos sites and upper size limit in the head of phage, the recombinant DNA molecules can be packed into bacteriophage particles in in vitro packaging system consisting of packaging enzymes, head and tail proteins.
Procedure of DNA cloning by using cosmid vector is shown in Fig. 3.8. Upon infection of E. coli by bacteriophage, the recombinant DNA cyclizes through cos sites and then replicates as a plasmid and expresses the drug resistance marker. Recently, based on cosmid vectors, a number of cosmid vectors have been determined from E. coli, yeast, and mammalian cells, and gene bank has been constructed (Dahl et al., 1981; Grosveld et al. 1982). Moreover, other vectors which have been used in cloning are bacteriophage M13, virus SV40, Ti-plasmid, etc. but no one of them are useful for building gene bank. Ti -plasmid and SV40 have been described elsewhere (see section on Animal viruses).

Fig. 3.8. Cloning of a cosmid vector. Transduced bacteria contain a cosmid which show resistance to specific antibiotic. Such bacteria can be screened. Antr, antibiotic resistance; ori, origin of replication.
Phasmids
The plasmids may be inserted into a phage λ genome, as a phage λ genome into the bacterial chromosome, during lysogenic cycle. Insertion of plasmid into phage λ is done with a view to have a specific site responsible for recombinational insertion of the phage into bacterial chromosome during lysogenic cycle. This insertion of plasmid into phage λ genome is reversible and referred to as 'lifting' the plasmid. It generates a phage genome containing att site and one or more plasmid molecule(s). These new genetic recombinations are called as phasmids (Brenner et al, 1982). The phasmids contain functional ori genes of plasmids and of phage λ. Moreover, they may be allowed to propagate as plasmid or phage in appropriate E. coli strains. Plasmids are released when reversal of lifting processes takes place.
Phasmids may be used in multifarious ways. They may be used as a phage cloning vector from which a recombinant plasmid may be released. Also phage particles are easy to store for a long time.