Fine Mapping of Gene Ordering by Elongated Chromosome Methods
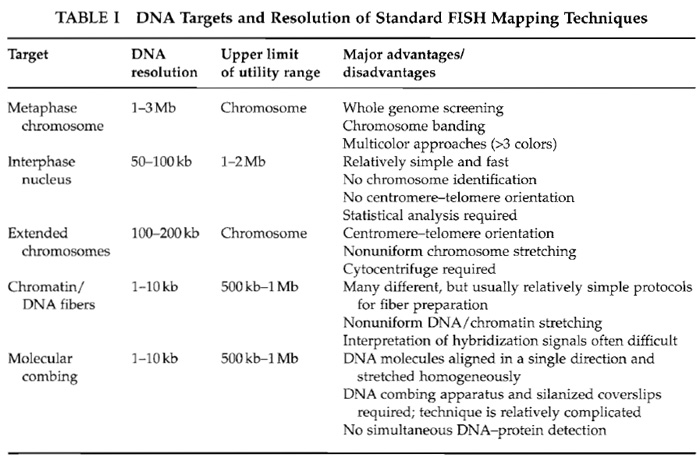
Fluorescence in situ hybridization (FISH) can be used to localize specific DNA sequences on metaphase chromosomes, interphase nuclei, and experimentally extended DNA or chromatin fibers. Depending on the hybridization target, FISH techniques show widely different levels of DNA resolution (Table I). They provide a rapid and accurate alternative to traditional molecular approaches for long-range physical mapping, including pulsed-field gel electrophoresis and radiation hybrid mapping. One important application has been the visual construction of contig maps of regions of interest, such as disease gene loci or chromosome breakpoints. Differential fluorescent probe labeling allows the ordering of multiple DNA sequences on the same chromosome. The DNA resolution of standard metaphase FISH is in the 1- to 3-Mb size range. Hybridization on less condensed interphase nuclei increases the resolution to around 100 kb. The average distance between two probe signals in the three-dimensional nuclear space correlates with physical distance up to 1-2Mb. This allows one to determine the relative order and proximity of closely juxtaposed sequences, which cannot be resolved on metaphase chromosomes (Trask et al., 1989). Protocols for stretching out DNA in solution (Bensimon et al., 1994) or the chromatin of interphase nuclei (Parra and Windle, 1993; Haaf and Ward, 1994b; Heiskanen et al., 1994) have pushed the limits of resolution even further. Hybridization to experimentally extended DNA or chromatin fibers provides a mapping power in the order of 1-10 kb. The maximum distance possible for clone ordering may be up to several megabases; however, the average size of intact fibers is considerably smaller. Most fiber FISH techniques are informative over genomic distances from 100 to 500 kb.
 |
Although various genome projects have provided very high-resolution physical maps of human and important animal genomes, FISH is still an exceptionally versatile mapping tool. For example, working with duplicated or repetitive sequences at the molecular level can be extremely tedious and, therefore, accurate mapping and sequencing of these regions remain tremendous tasks in the postgenomic era. In addition, FISH has important applications for mapping chromosome regions that have been deleted, amplified, and/or rearranged in human pathology or evolution. Of course, FISH techniques are useful not only for mapping the human genome. They can be adapted easily for the study of map-poor genomes from primitive vertebrates to humans.
All reagents for elongated chromosome preparation are standard laboratory chemicals that can be obtained from many different suppliers, i.e., NaCl (Sigma- Aldrich, Cat.No. S7653), KCl (Sigma-Aldrich, Cat.No. P5405), Na2HPO4 (Sigma-Aldrich, Cat.No. S9638), KH2HPO4 (Sigma-Aldrich, Cat.No. P5655), HEPES dry powder (Sigma-Aldrich, Cat.No. H9897), glycerol 99.5% ACS reagent (Sigma-Aldrich, Cat.No. G7893), CaCl2·2H2O (Sigma-Aldrich, Cat.No. C3881), MgCl2·6H2O (Sigma-Aldrich, Cat.No. M0250), acetic acid (VWR, Cat.No. 1.00058.1000), methanol (VWR, Cat.No. 1.06008.1000), acetone (VWR, Cat.No. 1.59005.0500), Biocoll (Ficoll) separating solution (Biochrom, Cat.No 2 6115), and colcemid (Roche, Cat.No. 295892). The cytocentrifuge (Cytospin 3) and the filter blots are from Thermo Shandon Inc.
III. PROCEDURES
A. Preparation of Elongated Chromosomes for FISH Mapping
The following protocol describes the preparation of elongated chromosomes from human lymphoblastoid or fibroblast cells (Haaf and Ward, 1994a,b). This technique can be adapted for other cell substrates, producing suitable targets for high-resolution FISH mapping.
Solutions
- Phosphate-buffered saline (PBS): 136mM NaCl, 2mM KCl, 10.6mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.3. To make 10× PBS stock solution, weigh 79.5g NaCl, 1.5 g KCl, 15 g Na2HPO4, and 2 g KH2PO4. After dissolving in distilled water, complete to 1000ml, pH, and autoclave. Store at room temperature. To make 1× PBS, dilute stock solution 1 : 10 with distilled water and use within 1 day.
- Hypotonic solution (for cell swelling): 10mM HEPES, 30mM glycerol, 1 mM CaCl2, and 0.8mM MgCI2. To make 1000 ml, add 2.4g HEPES, 2.2ml anhydrous glycerol, 147mg CaCl2·2H2O, and 163mg MgCl2·6H2O. After dissolving, complete to 1000ml with distilled water, pH, and filter if necessary. Store at 4°C and use within 1 week.
- Carnoy's fixative: Mix one part acetic acid and three parts methanol. Store at -20°C and use within 1 day.
- After harvesting a mitotically dividing cell culture(s), wash the cells one time in 1× PBS and resuspend the cell pellet in hypotonic solution to a concentration of 103 to 104 cells/ml. Incubate the cells at ambient temperature for 5-10min.
- Centrifuge 0.5-ml aliquots of this hypotonic cell suspension onto microscope slides at 1000rpm for 4min using a Shandon cytospin. Specifically, place the cell suspension in the cytobuckets of the centrifuge, the bases of which are formed by a filter blot and a glass slide (cleaned with ethanol). The whole structure is held together by a clip. During centrifugation the suspension fluid is absorbed by the filter blot and the cells become attached to the glass slide. After cytocentrifugation, the slide should carry a visible monolayer of cells in a designated area about 5 mm in diameter. Use a diamond pencil to mark the back of the slide to indicate the area on which the cells are sedimented.
- Fix the slides for 30min at -20°C in methanol and then overnight at -20°C in Carnoy's fixative. Airdried slides stored at 4°C can be kept for months before use.
- Select slides suitable for FISH mapping using a phase-contrast microscope at low magnification (10 to 20× lens without oil). A higher density of cells per slide results in poor chromosome stretching and cytoplasmic background ("dirt"). A lower density of cells results in disruption of metaphase spreads and individual chromosomes. Chromosomes extended to 5 to 20 times their normal length but still intact are most useful for high-resolution chromosome mapping. Good slides should contain 3-10 metaphase spreads with stretched chromosomes without broken arms. If less than half of the slides obtained after cytocentrifugation meet these criteria, the cell density in the hypotonic solution was either too high or too low.
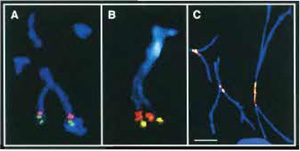
- Standard FISH protocols are used for mapping cloned DNA sequences such as cosmids, BACs, or YACs to acid-fixed elongated chromosomes (Haaf, 2000). Usually one probe is detected with a green fluorescent dye, a second probe with a red dye, and the chromosomes are counterstained in blue with DAPI (Figs. 1A and 1B). However, using modern FISH workstations for M-FISH or spectral karyotype analysis (SKY), many more DNA targets can be distinguished in different colors.
- Determine the relative order of two hybridized clones that are <1Mb apart on at least five elongated chromosomes. Because mechanically stretched chromosomes can be highly deformed, not all hybridized chromosome copies may be informative. If the order is not consistent on all five chromosomes, analyze five additional chromosome copies. In most cases, one slide should be sufficient for ordering probes that are 100-200 kb apart.
The combined application of protein staining by immunofluorescent techniques and DNA staining by FISH provides a powerful tool for mapping chromosomal proteins to specific DNA sequences. Steps
- Harvest mitotic cells and centrifuge them onto slides as described earlier.
- Fix the slides in methanol at -20°C for 30 min and then immerse in ice-cold acetone for a few seconds. After brief air drying, process the preparations for immunofluorescence staining. Because most protein antigens are not very stable, it is not recommened to store the slides before use.
- Perform immunofluorescent staining of a protein of interest, i.e., as an example only, labeling of centromeric proteins (CENPs) by specific anti-CENP antibodies (Fig. 1C). Standard immunofluorescent protocols are used for incubation of the slides with primary and fluorescent-labeled secondary antibodies (Haaf, 1995). However, do not counterstain the preparations with DAPI.
- Immediately after immunofluorescent staining, refix the preparations by incubating them in Carnoy's fixative overnight at -20°C and then hybridize them with a DNA probe(s) of interest.
- The most suitable source of mitotic cells are longterm in vitro cell cultures such as EBV-transformed lymphoblasts, fibroblasts, or established cell lines. Usually, 10µg/ml colcemid (Roche) is added to the culture medium 10-15 min before cell harvest to arrest cells in metaphase. Whole blood cultures cannot be used as a cell substrate because the erythrocytes are not destroyed by the hypotonic treatment required for elongated chromosome preparation. Therefore, it is necessary to separate white blood cells from red cells by density gradient centrifugation (in Biocoll/Ficoll separating solution) prior to short-term lymphocyte culturing.
- Elongated chromosomes constitute a compromise between different demands. The mechanical stretching procedure inevitably impairs chromosome morphology. The chromosomes are deformed and cannot be banded; however, they provide a greater DNA resolution and allow the ordering of probes from the short-arm to the long-arm telomere. Usually, better chromosome morphology goes along with poor stretching and vice versa.
- Chromosome elongation is not uniform over the entire length of a stretched chromosome and the same metaphase spread may contain both well-extended and poorly stretched chromosomes. Therefore it is very difficult to correlate the micrometer distance between two mapped loci or the relative distance from the short arm telomere (Flpter value) with physical distance in kilobases. Some regions, i.e., telomeres, which are usually attached to the surface of the glass slide, are more resistant to stretching than others.
- Carnoy's fixation of immunofluorescent preparations and subsequent FISH do not grossly interfere with the fluorecent-labeled protein antigen-DNA complex. Although the intensity of the fluorescent protein signal is reduced to a certain extent, it is still visible after the relatively harsh in situ hybridization procedure. This allows one to analyze the results of protein immunofluorescence and DNA in situ hybridization simultaneously on the same elongated chromosomes.
Bensimon, A., Simon, A., Chiffaudel, A., Croquette, V., Heslot, F., and Bensimon, D. (1994). Alignment and sensitive detection of DNA by a moving interface. Science 265, 2096-2098.
Haaf, T. (1995). Immunocytogenetic techniques. In "Human Chromosomes. Principles and Techniques" (R. S. Verma, A. Babu, eds.), pp. 232-270. McGraw-Hill, New York.
Haaf, T. (2000). Fluorescence in situ hybridization. In "Encyclopedia of Analytical Chemistry" (R. A. Meyers, ed.), Vol. 1, pp. 4984-5006. Wiley, Chichester.
Haaf, T., and Ward, D. C. (1994a). High resolution ordering of YAC contigs using extended chromatin and chromosomes. Hum. Mol. Genet. 3, 629-633.
Haaf, T., and Ward, D. C. (1994b). Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 3, 697-709.
Heiskanen, M., Karhu, R., Hellsten, E., Peltonen, L., Kallioniemi, O. P., and Palotie, A. (1994). High resolution mapping using fluorescence in situ hybridization to extended DNA fibers prepared from agarose-embedded cells. BioTechniques 17, 928-933.
Laan, M., Isosomppi, J., Klockers, T., Peltonen, L., and Palotie, A. (1996). Utilization of FISH in positional cloning: an example on 13q22. Genome Res. 6, 1002-1012.
Parra, I., and Windle, B. (1993). High resolution visual mapping of stretched DNA by fluorescent hybridization. Nature Genet. 5, 17-21.
Trask, B., Pinkel, D., and van den Engh, G. (1989). The proximity of DNA sequences in interphase nuclei is correlated to genomic distances and permits ordering of cosmids spanning 250 kilobase pairs. Genomics 5, 710-717.