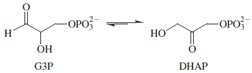Triosephosphate Isomerase
Triosephosphate isomerase (TIM) catalyzes the interconversion
of D-glyceraldehyde-3-phosphate (G3P) and
dihydroxyacetone phosphate (DHAP). The equilibrium
lies far to the side of DHAP, hence the longer arrow
pointing to that compound. The enzyme operates with
a turnover number of ∼10
7 s
−1, which is nearly as fast
as the diffusion-controlled
limit. TIM is therefore called
an almost perfectly evolved enzyme because no catalytic
refinement could make the rate faster than it already is.
Many tools have been used to study the TIM mechanism,
including X-ray crystallography and NMR,
site-directed mutagenesis, and affinity labeling. Strong
evidence for the mechanism, however, was supplied by
studies using isotopic labeling of substrates. It was found
that if the above reaction was carried out in tritiated water,
one atom of tritium was stereospecifically incorporated
into DHAP.
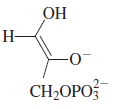
This result suggested that a base abstracts a proton from
the substrate, and the proton then undergoes exchange with
labeled protons from the solvent before being added back
to form the
product stereospecifically. The existence of a
cis-enediol intermediate (shown below)would account for
these observations, if the enzyme added the proton back
to the same face of the enediol that it was abstracted from.
One unresolved questionwas whether only a single protein
base was involved (so that the transfer was from substrate
to base and directly back to form product) or whether
a different base was responsible for protonation as part of
a more extensive proton relay. The nature of the protein
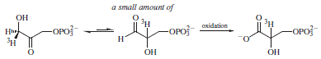
base was explored by doing a similar experiment to the
one described above but in the other direction; that is, by
labeling the DHAP and observing its conversion to G3P.
Although the equilibrium lies far to the side of the DHAP,
trapping by irreversible oxidation by G3P dehydrogenase
of any G3P formed was used to convert significant quantities
of DHAP. If the DHAP was labeled at C1, a small
but measurable amount of the label was transferred to C2.
 |
This result suggested that a single base was involved in
the proton abstraction/proton addition step. If more than
one base were involved, the chance that any label would
not be washed out by the solvent and would be added
to the deprotonated intermediate would be vanishingly
small. In combination with other kinds of experiments,
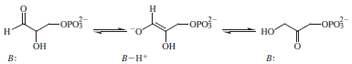
isotopic labeling was therefore invaluable in elucidating
the mechanism of triosephosphate isomerase (shown below)
in which
B is a protein-derived base.
Isotopes can be used in another way to measure the
energy barrier heights for various steps in the catalytic
mechanism as noted above for the reaction catalyzed by
dihydrofolate reductase. For example, if a proton transfer
is involved in the rate-limiting step, then substitution of
that proton with one of the heavier isotopes of hydrogen
(deuterium or tritium) will cause the step to proceed more
slowly. These so-called kinetic isotope effect experiments
in combination with steady-state rate measurements in the
case of TIM allowed the elucidation of the rate constants
for partitioning of the cis-enediol intermediate and construction
of a detailed kinetic scheme as shown above for
dihydrofolate reductase.