The experimental approaches for the study of developmental biology in higher plants range over the fields of morphology, physiology, biochemistry and molecular biology. The genetic approach, coupled with the techniques of biochemistry and molecular biology offers unparalleled opportunities for the study of molecular basis of plant development.
Mutations that effect development of flower primordia (class I mutants). Sterilis and
steriloides are two mutants, where abnormal inflorescences carrying only bracts, but no flowers are produced, although occasionally few deformed flowers are produced in
steriloides mutants. This suggests that these genes are involved in the development of flower primordia.
Homeotic genes in flower development. Although several plant species have been used for a study of the genetics of development, the most advanced study pertains to the molecular and genetic analysis of floral homeotic genes in
Arabidopsis thaliana and
Antirrhinum majus. The use of
Arabidopsis as an experimental material has the following advantages : (i) small genome size, (ii) small duration of generation time, (iii) ease of transformation and (iv) small plant volume.
Similarly, the use of
Antirrhinim has the following advantages : (i) well characterized transposable elements, (ii) large flowers, (iii) easy to emasculate and cross and (iv) ease of vegetative propagation.
(a) Genetic control of first steps. A large number of mutants for flower development have been isolated, which could be broadly classified into two groups, (i)
homeotic mutations that cause the transformation of one flower whorl into a whorl of a different type (e.g.
apetala, where petals are absent and converted in bracts, stamens or carpels) and (ii)
meristic mutations that alter the number of a structure within a floral whorl (e.g.
clavata, where number of carpels increased from two to three or four). A number of mutations including both these types, known in
Arabidopsis are listed in Table 38.1. Analogous mutations have been observed in other plant species, indicating that the fundamental processes controlling floral morphogenesis are highly conserved. In wild type plants, after a period of vegetative growth,
the apex undergoes a transition to become an inflorescence meristem to produce indeterminate inflorescences, which do not produce a terminal flower. The inflorescence bears bracts, in the axils of which flowers develop. Two classes of mutants showing deviation from wild type have been detected in
Antirrhinum : (i)
floricaula (
fid)mutation '(called
leafy in
Arabidopsis)leads to failure in transition from inflorescence meristem to floral meristem, so that proliferating inflorescence shoots develop in place of flowers. Two other mutations
squemata and
squamosa giving phenotypes similar to that of
floricaula (
flo)were reported, (ii)
centroradialis mutation has an effect opposite to that
of flo and leads to the development of terminal flower converting indeterminate inflorescences into determinate type. Mutations producing proliferating inflorescences have also been obtained in alalfa, tomato, maize, etc.
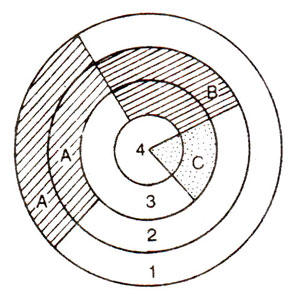
Fig. 38.6. Three regions of floral meristem, i.e., A, B and C, such that genes (a, b and c) acting in these regions have regulatory functions; note that out of four whorls (1 to 4), whorl 1 is controlled by a alone; whorl 2 is controlled by a and b; whorl 3 is controlled by b and c and whorl 4 is controlled by c alone.
The
flo gene has been isolated and was shown to produce a transcript of 1.6kb, encoding a protein FL0 of 396 amino acids showing some similarity with transcription factors (see
Expression of Gene : Protein Synthesis 2. Transcription in Prokaryotes and Eukaryotes).
In situ hybridization shows that
flo is expressed from a very early stage in wild type inflorescence in a very specific temporal and spatial sequence. It expresses in bract primordia followed by expression in primordia for sep.als, petals and carpels but not in the primordia for stamens. Therefore,
flo+ may be involved in transition from vegetative to floral axis and in the initiation of floral primordia. Absence of its expression, however, is required for normal stamen development. The
flo+ may also interact with other genes, like those meant for whorl identity.
(b) Mutations that alter the symmetry of the flower (
class II mutants). Mutations that change zygomorphic flowers into actinomorphic or
vice versa have also been identified. In
Antirrhinum, cycloidia and
centroradialis are two mutants, both conferring radial symmetry (actinomorphic flowers) on otherwise zygomorphic flowers. As reported above,
cantroradialis gives radial symmetry due to the terminal position of the flowers, which are normally developed only in the axils of bracts.
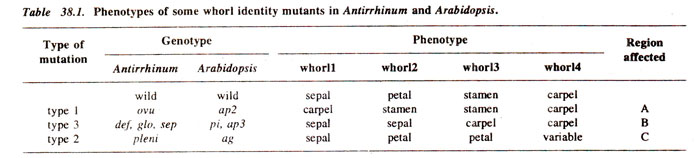
(c) Genetic control of whorl identity (class III mutants).A major class of whorl identity genes (which will affect the identity of organs in a whorl) contains those that affect the identities of organs in two adjacent whorls (Table 38.1). For convenience, the following three overlapping regions, each consisting of two adjacent whorls, were recognized: A (whorls 1 and 2), B (whorls 2 and 3) and C (whorls 3 and 4) (see Fig. 38.6). Consequently, as shown in Table 1, three classes of mutants (called homeotic mutants) are known, (i) those affecting whorls 1 and 2 (type 1); (ii) those affecting whorls 2 and 3 (type 3) and (iii) those affecting whorls 3 and 4 (type 2). The genes acting in these three regions (A, B and C) are required for three functions
a, b and
c.
Experiments on isolation and cloning of homeotic genes revealed that many homeotic genes encode transcription factors. For instance
def gene encodes DEFA protein which resembles transcription factors like serum response factor (SRF) in mammals. Similarly, the product of minichromosome maintenance gene (MCM 1) in yeast, has been shown to be a transcription factor. Similar is the situation with the
agamous (
ag)gene in
Arabidopsis producing a product AG. These genes seem to have a conserved domain in common with each other and include the gene products MCM 1, AG, DEF A and SRF. Therefore, this domain has been given the name MADS-box.

Fig. 38.6. Three regions of floral meristem, i.e., A, B and C, such that genes (a, b and c) acting in these regions have regulatory functions; note that out of four whorls (1 to 4), whorl 1 is controlled by a alone; whorl 2 is controlled by a and b; whorl 3 is controlled by b and c and whorl 4 is controlled by c alone.
The study of the phenotypes in several mutants as above followed by a study of cloned genes led several workers to formulate models for floral development. But the information is inadequate yet to suggest complete models based on unequivocal evidence. Knowledge about regulation of the expression of these genes is being generated at a rate, which should permit formulation of better models for the understanding of the pathways for flower development.