Although all enzymes needed for recombination have not been identified, enzymes involved in some specialized recombination events have been characterized. RecA protein and RecBCD endonuclease have been purified from bacterial systems, utilizing Rec
- mutants, which are unable to undertake generalized recombination. However, there may be a number of other products that may be involved in recombination process, but could not be isolated. Topoisomerases are another class of enzymes involved in recombination. Phage lambda (λ)also utilizes
Int protein, Xis protein and
IHF protein for recombination.
Rec A and Rec BCD proteins promote base pairing between a single strand of DNA and its complement available in double stranded DNA molecule. The single strand displaces its homologue (strand which is similar to the invading single strand) in the duplex and base pairs with its complement through a reaction described as
single-strand uptake or
single-strand assimilation.
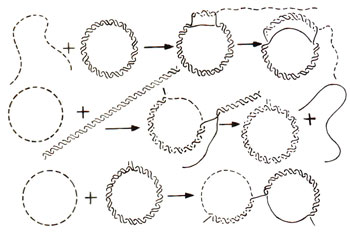
Fig. 13.9. Different forms of reaction for invasion of single strand into a duplex, with the help of Rec A.
The single stranded region, however, must have a free 3' end and a segment complementary to one of the two strands of the duplex. Some forms of this reaction are shown in Figure 13.9. It is believed that the Rec A protein first binds to single stranded DNA and then binds double stranded DNA and looks for the region complementary to single stranded DNA. This single-strand assimilation seems to be related to the initiation of recombination. The models for recombination earlier described in this section, involve an intermediate stage, where both single strands cross over from one duplex to the other. Rec A protein may presumably catalyze this reaction, even though such a protein has not so far been isolated from a eukaryotic system.

Fig. 13.9. Different forms of reaction for invasion of single strand into a duplex, with the help of Rec A.
There are some other attributes of Rec A, which make its role in recombination understandable. These include the following : (i) ability to polymerize single strands; (ii) ability to incorporate duplex DNA into filaments, giving duplexes longer than the original duplex. In view of this, the reaction involving generation of heteroduplexes during recombination may involve following three steps : (i) a slow presynaptic phase, in which Rec A polymerizes on single stranded DNA; (ii) a fast pairing reaction between single stranded DNA and its complement in double stranded DNA, to produce heteroduplex DNA; (iii) a slow displacement of one strand from the duplex, to produce a long region of heteroduplex DNA. The initial region of heteroduplex DNA may not be found in a double helical form
(plectonemic), but the two strands may be associated side by side
(paranemic joint). This paranemic joint is unstable and further progress in recombination events requires its conversion to double helical form.
When the reacting DNA molecules are circular, the product of Rec A sponsored reaction is a chi (χ) structure. Although action of Rec A is independent of DNA sequence, certain hot spots stimulate the Rec A recombination system. These hot spots were discovered in lambda phage
chi mutants and share common non-symmetrical sequence of 8 bp (5'GCT GG TGG3'). This sequence is called
chi sequence and is not necessary for recombination. However, it stimulates recombination in its vicinity and identifies targets for the enzyme exonuclease V synthesized under the control of
Rec BCD genes. This enzyme has following roles : (i) it degrades DNA; (ii) it can unwind duplex
DNA in the presence of SSB; (iii) it has an ATPase activity. Thus it provides a single stranded region with a free end. These free ends are then seized by Rec A protein, which requires free termini to start the strand assimilation reaction.
Topoisomerases. Recombination, like replication, also requires topological manipulation of DNA, which is facilitated by topoisomerases. These enzymes may relax or introduce supercoils in DNA and are required to disentangle DNA molecules that have become catenated by recombination or replication. Topoisomerases can be of
Type I (making transient break in one strand) or
Type II (making transient double strand breaks). Type I topoisomerases (e.g. product of
top A gene of
E. coli)relax negatively supercoiled DNA, while Type II topoisomerases (e.g.
gyrase) relax both negative and positive supercoils, (for supercoiling consult
Chemistry of the Gene 1. Nucleic Acids and Their Structure). The removal of supercoils from the crossing segments is required during the recombination process.






