Several
hybrid DNA or
heteroduplex DNA models to explain the mechanism of recombination have been proposed which assume breaks in only one of two strands of each of the two DNA molecules, belonging to two non-sister chromatids. Two such models were suggested, one each by
H.L.K. Whitehouse and
Robin Holliday, the major difference in these two models being in the polarity of the two strands undergoing single breaks.
In the model proposed by Whitehouse, these two strands will have opposite polarity (5'→3' and 3'→ 5'), but in Holliday's model, their polarity will be similar. Since Holliday's model received wide support and has been variously modified and discussed, this will be briefly described here. The model has been widely accepted both for eukaryotes and prokaryotes.

Fig. 13.1. The phenomena of branch migration, which can occur in either direction, when an unpaired single strand displaces a the paired strand.
A paired bivalent in meiosis consists of four chromatids and at a particular location, two of them take part in recombination, although for double and : multiple crossovers, three or all the four chromatids may be involved. The process starts with breakage, at the corresponding points, of the homologous strands of two paired DNA duplexes, each representing a chromatid, the other two chromatids being not involved.
The breakage allows movement of the free ends created by the nicks. Each strand at the broken end leaves its partner, crosses over broken strand of the other non-sister molecule and pairs with its complement in the other DNA duplex (by a phenomenon called
branch migration; see Fig. 13.1.). It creates a connection between the two DNA duplexes. Initially this connection is sustained by hydrogen bonding, but at some point during branch migration, the nicks at the sites of exchange are sealed with the help of ligase enzyme. The connected pair of duplexes is called a
joint molecule and the point at which an individual strand of DNA crosses from one duplex to the other is called the
reconibinant joint. At the site of recombination, each duplex has a region consisting of one strand from each of the two parental DNA molecules. This region is called
hybrid DNA or
heteroduplex DNA.

Fig. 13.1. The phenomena of branch migration, which can occur in either direction, when an unpaired single strand displaces a the paired strand.
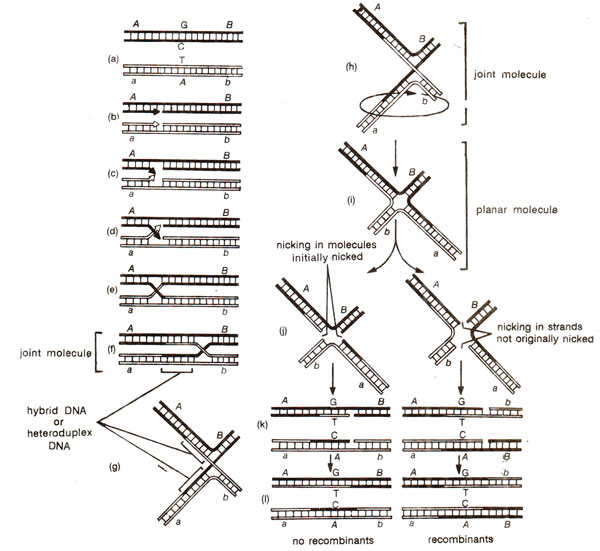
Fig. 13.2. A hybrid DNA model for the molecular mechanism of recombination; the recombination between two paired duplex DNAs could be initiated by reciprocal single strand exchange, (after single strand breaks), which is extended by branch migration and resolved by nicking.
The
joint molecule formed due to exchange of broken strands, needs, to be resolved from each other into two separate DNA molecules and this is possible only through a further pair of nicks. This is achieved in two steps : (i) One of the two DNA duplexes in the joint molecule undergoes rotation, giving rise to a
planar molecule, (ii) In the planar molecule, there are four strands and two of them are nicked. The consequences of nicking will depend upon, which pair of strands is nicked. Following two alternatives are possible : (i) If the nicks are made in the pair of strands that are not originally nicked (so that all the four strands are nicked now),
this leads to release of recombinant DNA molecules. The DNA duplex segment of one parent is covalently linked to the DNA duplex segment of the other parent
via a stretch of heteroduplex DNA. This means that conventional recombination has taken place between markers located on either side of the heteroduplex region, (ii) If the two strands involved in the original nicking are nicked again, then only original parental duplexes with heteroduplex segments, but with no recombinant DNA molecules, are released. Different steps envisaged in Holiday's hybrid. DNA model of recombination are shown in Figure 13.2.

Fig. 13.2. A hybrid DNA model for the molecular mechanism of recombination; the recombination between two paired duplex DNAs could be initiated by reciprocal single strand exchange, (after single strand breaks), which is extended by branch migration and resolved by nicking.








