Properties of genetic code
Following properties of the genetic code were proved by definite experimental evidence : (i) the code is triplet, (ii) the code is degenerate, (iii) the code is non-overlapping, (iv) the code is commaless, (v) the code is non-ambiguous and (vi) the code is universal. Although it may not be necessary to present the experimental evidences which proved the validity of these properties, it may be useful to explain the meaning of these six properties of the genetic code listed above.
The code is triplet
As earlier outlined, singlet and doublet codes are not enough to code for 20 amino acids; it was pointed out that triplet code is the minimum required. But it could be a quadruplet code or of a higher order. As pointed out above, in a triplet code of 64 codons, there is an excess of 44 codons and, therefore, more than one codons are present for the same amino acid. This excess will be still greater if more than three-letter words are used. In the quadruplet code there will be 4x4x4x4 = 256 possible words.
In a triplet code, as pointed out earlier also, for a particular amino acid more than cine words (synonyms) can be used. This phenomenon is described by saying that the code is degenerate. A non-degenerate code would be one where there is one to one relationship between amino acids and the codons, so that 44 codons out of 64, will be useless or nonsense codons. It has been definitely shown that there are no nonsense codons. The codons which were earlier called nonsense codons are also now known to mean stop signals.
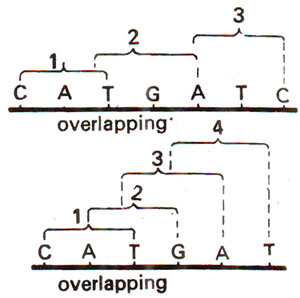
Fig. 30.2. Overlapping of codons due to one letter or two letters.
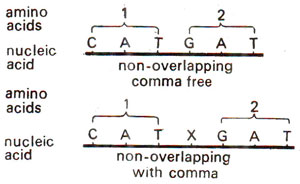
Fig. 30.3. Genetic code, without comma and with comma.
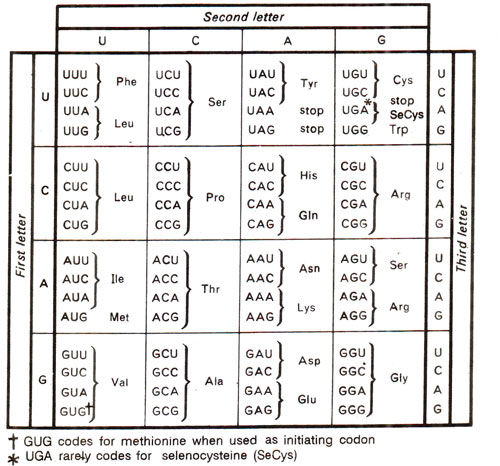
Fig. 30.4. Genetic code dictionary.
Non-overlapping code means that a base in a mRNA is not used for two different codons. In Figure 30.2 it is shown that an overlapping code can mean coding for four amino acids from six bases. In actual practice six bases code for not more than two amino acids. However, in overlapping genes described in Organization of Genetic Material 3. Split Genes, Overlapping Genes and Pseudogenes, it is shown that the same base can be used for different codons, but only at different occasions in time and/or space, so that the same base can not be used for two different codons during synthesis of the same protein. This is exactly, what we mean by non-overlapping code.
A commaless code means that no punctuations are needed between any two words. In other words, we can say that after one amino acid is coded, the second amino acid will be automatically coded by the next three letters and that no letters are wasted for telling that one amino acid has been coded and that now second should be coded (Fig. 30.3).
The code is non-ambiguous
Non-ambiguous code means that there is no ambiguity about a particular codon. A particular codon will always code for the same amino acid, wherever it is found. In an ambiguous code, the same codon could have different meanings, or in other words, the same codon could code two or more than two different amino acids. Such is not the case.
Although the code has been worked out by using in vitro systems prepared usually from micro-organisms, there is no doubt now that in all kinds of living organisms, micro or macro, plants or animals, the same genetic code is used. However, as will be seen later in this section, a different and more primitive genetic code exists in mitochondria of some organisms.
Codon Assignments
Although theoretical considerations in 1950's had suggested that the genetic code should be triplet in nature, it was not possible to say which codon of the possible 64 codons should code for which of the 20 essential amino acids. The first clue to this problem came when M.W. Nirenberg (Nobel Prize winner with H.G. Khorana and R.W. Holley in 1968) used in vitro system for the synthesis of a polypeptide using an artificially synthesized mRNA molecule. The use of artificial mRNA molecule has a definite advantage, since one could know something about its structure.
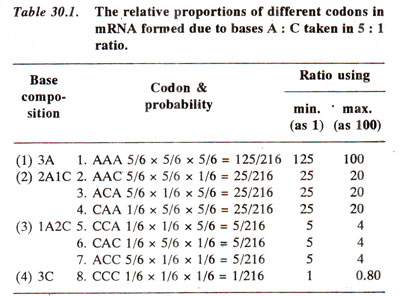
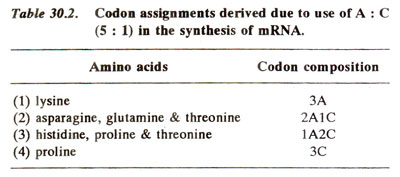
A large number of synthetic copolymers of unknown sequences was used for preparing a dictionary where codon compositions were assigned to amino acids. The next problem and a serious one was to sort out the three codons in one base composition e.g. 2A1C or 1A2C. In other words, we can say that the next problem was to work out the sequences of bases in codons, whose composition was already known. This was done mainly by the following methods.
Binding technique of Nirenberg and Leder. M.W. Nirenberg and P. Leder in 1964 found that if a synthetic trinucleotide for a known sequence (with known bases at 5' end and 3' end) is used with ribosome and a particular aminoacyl-tRNA (tRNA having its own specific amino acid attached), these will form a complex, provided the used codon codes for the amino acid attached to the given aminoacyl tRNA.
Codon1 + Ribosome + AA1-tRNA → Ribosome-Codon1-AA1 –tRNA1
It was also observed that while the free AA-tRNA passes through nitrocellulose membrane, the ribosome-codon-AA-tRNA complex adsorbs on such a membrane. If in a particular mixture only one of the amino acids is made radioactive, then the presence or absence of the radioactivity on the nitrocellulose membrane will show whether there is a relationship between the codon and the amino acid which was made radioactive.
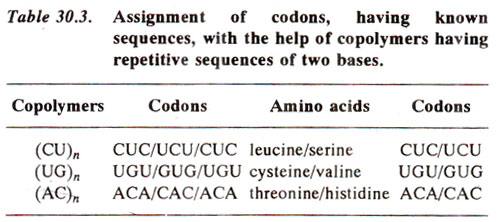
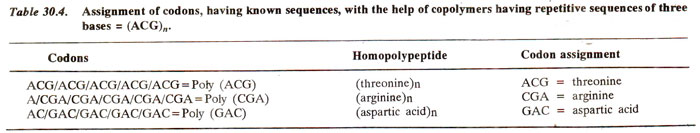
Copolymers of repetitive sequences by H.G. Khorana. As outlined above, sufficient information regarding the sequences of bases in codons was available through the painstaking work of Nirenberg and co-workers. However, H.G. Khorana, almost at the same time, devised an ingenious technique for the same purpose. Using synthetic DNA, Khorana and his co-workers could prepare polyribonucleotides (RNA) with known repeating sequences. A repeating sequence means that, if CU are two bases, these will be repeatedly present throughout the length as follows : CUCUCUCUCUCUCU
In a similar manner, if ACU are three bases they will be present repeatedly as follows : ACUACUACUACU