Suppressor mutations, base substitutions and suppressor tRNAs
When the effect of one mutation is suppressed by another mutation, the latter is called a suppressor mutation. For instance,
AABB may be wild, and
aaBB a mutant; then if
aabb, is also a wild type, the mutation in 'B' will be called suppression mutation, because the effect of
‘a’ is suppressed by
‘b’ Mechanism of this suppression effect was not known for some time, but now its mechanism is largely known. These effects can be conveniently classified into (i) intragenic suppression, and (ii) intergenic suppression.
Intragenic suppression
Suppression of a mutation at one site within a gene can be achieved by mutation at another site within the same gene. For instance, a mutant A46 that produces inactive
tryptophan synthetase enzyme, due to substitution of glutamic acid for glycine,
at position 210 may be corrected or suppressed due to mutation A446 involving substitution of cysteine in place of tyrosine at position 174. These two mutants A46 and A446 independently give rise to inactive enzyme, but if present together give rise to active enzyme, thus suppressing the mutant effect of each other. In many other combinations, double mutants do not show any suppression effect. The suppression effect really depends on whether or not the second alteration, can restore the 3-D configuration of the enzyme. There are a number of known examples of such intragenic or intracistronic suppression or complementation. The reader is advised to consult an advanced book for further details in this connection.
Intergenic suppression
Intergenic suppression, which means suppression of mutation in one gene due to mutation in another gene, can be achieved mainly due to alteration in transfer RNA (tRNA). The suppression may also be effective by alteration brought about in ribosome conformation. There are atleast four types of such suppression mutations, which can be discussed.
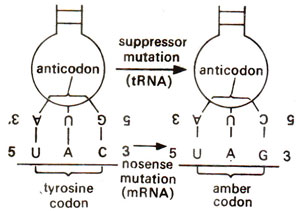
Fig. 30.9. Suppressor mutation involving a change in one base of anticodon of tyrosine tRNA leading to reading of termination amber codon UAG, which resulted from nonsense mutation in tyrosine codon UAC thus leading to suppression.
Nonsense suppression and mutant tRNA. Sometimes due to a change in anticodon of a specific tRNA, it may develop the capacity to read the mutant terminating codon
(amber or UAG,
ochre or UAA and
opal or UGA mutants). Since these nonsense mutations cause premature termination of polypeptide chain, they can be suppressed if a mutant tRNA can read the terminating codon and substitute an amino acid at the location of termination codon. Mutant tRNAs are known which are capable of recognizing termination codons and insert amino acids like
tyrosine, serine or
glutamine to restore the function of protein (Fig. 30.9). However these mutant tRNAs may also read the normal stop signals of proteins thus causing elongation of protein beyond its normal termination thus causing disturbances. To prevent this disturbance, double stop signals are often available in mRNA, so that if mutant tRNA reads one, the other may still bring about termination. Moreover, two genes may code for same tRNA, so that if one (minor gene) mutates, and gives rise to suppressor tRNA the other (major gene) still functions and gives rise to normal tRNA.

Fig. 30.9. Suppressor mutation involving a change in one base of anticodon of tyrosine tRNA leading to reading of termination amber codon UAG, which resulted from nonsense mutation in tyrosine codon UAC thus leading to suppression.
Transfer RNA (tRNA) mediated UAG suppression was first observed, when in coat protein of phage R 17, amber codon 5'UAG3' could be read by a tyrosine suppresser tRNA (Fig. 30.9), in which the anticodon had mutated from 5'GUA3' to 5'CUA3'. There are also other examples of this type of suppression.
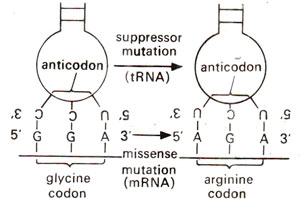
Fig. 30.10. Suppressor mutation involving a change in one base of anticodon of glycine tRNA leading to misreading of arginine codon AGA (as glycine) which resulted due to missense mutation in glycine codon GGA thus leading to suppression.
Missense suppression and mutant tRNA. We have already discussed mutations resulting due to base substitutions leading to replacement of one amino acid by another (see
Mutations: 3. Molecular Level (Mechanism)). These are called missense mutations. These mutations will lead to synthesis of a protein with a wrong amino acid at a particular site. Such a mutation can also be suppressed like nonsense mutations, by mutant tRNA, which will misread the codon and thus substitute a correct amino acid, although the mutant codon codes for a different amino acid. For instance, in mutant A36 in
tryptophan synthetase of
E. coli, glycine (coded by GGA) is replaced by arginine (coded by AGA). A suppresser mutant may have tRNA, depositing glycine for codon AGA actually meant for arginine (Fig. 30.10).

Fig. 30.10. Suppressor mutation involving a change in one base of anticodon of glycine tRNA leading to misreading of arginine codon AGA (as glycine) which resulted due to missense mutation in glycine codon GGA thus leading to suppression.
Frameshift mutations and mutant tRNA. In
Mutations: 3. Molecular Level (Mechanism), a reference was made about frameshift mutations. Earlier in this section these mutations were discussed, where the reading frame of mRNA shifts to the right due to deletion or to the left due to addition of a base pair in DNA. It has been shown that a frameshift mutation due to a deletion can be suppressed by mutant tRNA having an anticodon of two bases rather than three (doublet anticodon rather than triplet). Similarly, addition frameshift mutations can be suppressed by mutant tRNAs each having a quadruplet anticodon instead of a triplet. Mutant tRNAs with doublet and quadruplet anticodons have actually been detected in biological systems.
Suppression and ribosomal mutations. Mutations in some ribosomal proteins may cause a change in ribosome structure, so that it may now misread the mRNA codons. One such mutation, called
ram (ribosomal ambiguity) is capable of reading all the three termination codons, thus causing suppression of nonsense mutations. Some frameshift mutations may also be suppressed by these
ram mutants.
Mutations may also occur in ribosomes rendering them sensitive to an antibiotic like streptomycin. These sensitive ribosomes may then allow misreading of mRNA codons, although in resistant ribosomes this misreading is much less frequent. For instance, when Poly-U is used as a template with sensitive ribosomes, most frequent error is replacement of phenylalanine (coded by UUU) by isoleucine (coded by AUU). It has been shown that streptomycin sensitive strains carry mutations in 30S-ribosomal proteins, causing suppression through misreading.








