Mutant Analysis Allowed Identification of Genes for Cellulose Synthases and Other Proteins Required for Cellulose Biosynthesis
Identification and Functional Characterization of Cellulose Synthases in
Plants by Analysis of Mutants and Gene Expression Studies
Although a majority of the
CesA and
Csl genes have been identified from
genome and EST sequences, at least six of the
CesA genes in
Arabidopsis were
identified by mutant analysis. In a number of cellulose-deficient
Arabidopsis mutants, the mutations were mapped to genes that encoded for cellulose synthases (Arioli
et al., 1998; Fagard
et al., 2000; Scheible
et al., 2001; Taylor
et al.,
1999). Interestingly, although all the mutants exhibited different phenotypes, they
all showed a deficiency in the amount of cellulose produced. The first mutant,
where the mutation was identified in a gene that encoded for a cellulose synthase,
was a temperature-sensitive root-swelling mutant (
rsw1) (Arioli
et al., 1998). At the
nonpermissive temperature, the mutant produced a larger proportion of noncrystalline
cellulose in place of crystalline cellulose, and the rosette terminal complexes
(TCs) normally associated with cellulose microfibrils were not observed by
freeze-fracture electron microscopy. The mutation in the cellulose synthase gene
(
rsw1 gene; At
CesA1) led to the substitution of valine for alanine at position 549 of
the cellulose synthase protein and this change resulted in all the different phenotypes
associated with the
rsw1 mutant (Williamson
et al., 2001).
No biochemical
changes have been characterized in the mutant protein, but it appears that at
the nonpermissive temperature, the cellulose synthase is not assembled into a
rosette structure. Although the mutation results in the reduction of crystalline
cellulose at the nonpermissive temperature, noncrystalline cellulose still is
produced suggesting that the
rsw1-encoded cellulose synthase is able to synthesize
the β-1,4-glucan chains, but does not allow for their assembly to take place, or
alternatively these chains are synthesized by cellulose synthases encoded by other
genes, where the assembly of these cellulose synthases is affected by the
rsw1 mutation. Changes in cell shapes and sizes suggested that the
rsw1 cellulose
synthase contributed to cellulose in the primary wall. Interestingly, a number of
questions still remain to be answered in terms of how the
rsw1 mutation affects
cellulose biosynthesis.
A number of
irregular xylem mutants (
irx mutants) have been isolated by
screening cross-sections of stems of
Arabidopsis plants (Turner and Somerville,
1997). The mutations resulted in collapse of mature xylem cells in the inflorescence
stems, and in many of these mutants there was a significant decrease in the
amount of cellulose in the secondary cell wall of cells in the xylem. Genes mutated
in some of the
irx mutants were identified to encode for cellulose synthases.
The
null mutation in the
irx3 mutant results in a stop codon that truncates the cellulose
synthase (
irx3; At
CesA7) by 168 amino acids (Taylor
et al., 1999) In two
irx1
mutants (
irx1-1 and
irx1-2), the mutations were mapped to a different cellulose
synthase gene that altered the amino acids at positions 683 (D
683N) in
irx1-1 and
679 (S
679L) in
irx1-2 (Taylor
et al., 2000). Both these amino acid positions reside within the conserved region of the
irx1 cellulose synthase (At
CesA8). RNA analysis
indicated that
irx1 and
irx3 are highly expressed in stems but not in leaves,
suggesting that both genes are involved in cellulose synthesis during secondary
cell wall formation. Examination of the phenotypes of the xylem elements by
electron microscopy showed that the same cell type is affected in the
irx1 and
irx3 mutants, indicating that products of both the
irx1 and
irx3 genes are required
within the same cell for normal cellulose synthesis during secondary cell wall
formation (Taylor
et al., 2000). These results allowed development of the concept
regarding the nonredundant nature of cellulose synthases and the requirement of
more than a single cellulose synthase in each cell for normal cellulose synthesis.
Using biochemical and immunological methods, Taylor
et al. (2000) furthermore demonstrated that the
irx1 and
irx3 cellulose synthases associate with each other,
and suggested that this association is required for cellulose synthesis (Taylor
et al.,
2000).
Even as different models to explain the requirement of two different
cellulose synthases for cellulose synthesis were being proposed, another gene
(
irx5) encoding for a different cellulose synthase (
irx5; At
CesA4) was identified
in a further screen of
irx mutants and it was found that the
irx1,
irx3, and
irx5
genes were coexpressed in the same cells (Perrin, 2001; Taylor
et al., 2003). Using
detergent-solubilized extracts, the proteins encoded by these three genes were
shown to interact with each other, and it was suggested that all three gene
products probably are required for the formation of the cellulose-synthesizing
complexes (rosette TCs) in plants. Interestingly, the presence of all three cellulose
synthases (At
CesA8, At
CesA7, and At
CesA4), but not their activity, is required
for correct assembly and targeting of the cellulose-synthesizing complex during
secondary wall cellulose synthesis (Taylor
et al., 2004). Overall, the
irx mutants
have been crucial in not only identifying the cellulose synthase genes that are
required for cellulose synthesis during secondary wall formation, but also in
formulating the concept that the assembly of the cellulose-synthesizing complexes
(rosette TCs) in plants requires more than a single isoform of cellulose synthase.
Fig. 6.3 shows immunogold labeling of the rosette TCs from
Vigna angularis using
an antibody to a cellulose synthase.
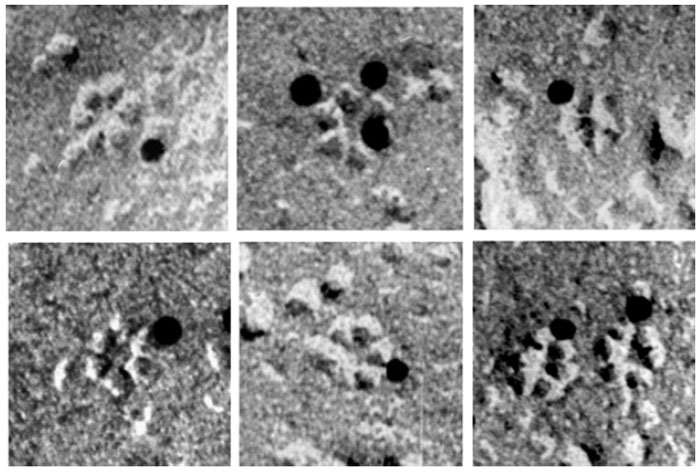 |
| FIGURE 6.3 Rosette terminal complexes from V. angularis that were immunogold labeled with
an antibody to cellulose synthase. (Reproduced from Kimura, S., Laosinchai, W., Itoh, T., Cui, X.,
Linder, R., and Brown, R. M., Jr. (1999). Plant Cell 11, 2075–2085.) |
The protein regulator of cytokinesis 1 (PRC1) gene in
Arabidopsis encodes
At
CesA6, and like the
rsw1 mutant of At
CesA1, mutation in this gene exhibits
decreased cell elongation, especially in roots and dark-grown hypocotyls, because of cellulose deficiency in the primary wall (Fagard
et al., 2000). In addition to
similar mutant phenotypes, both At
CesA1 and At
CesA6 also show similar expression
profiles in various organs and growth conditions suggesting coordinated
expression of at least two distinct cellulose synthases (At
CesA1 and At
CesA6) in
most cells (Fagard
et al., 2000). However, differences were observed in the embryonic
expression of these two
CesA genes (Beeckman
et al., 2002). Mutations in the
ixr1 and ixr2 genes confer resistance to the cellulose synthesis inhibitor isoxaben
and these two genes encode At
CesA3 and At
CesA6, respectively (Desprez
et al.,
2002; Scheible
et al., 2001). The cellulose synthases identified by analysis of the
rsw1, ixr1, and PRC1/ixr2 mutants involve members of the
CesA family (At
CesA1,
At
CesA3, and At
CesA6) required for primary wall cellulose synthesis. Although
no physical interactions have been determined for these cellulose synthases,
studies of inhibition of cellulose synthesis by isoxaben suggest that At
CesA3
and At
CesA6 together form an active protein complex in which the involvement
of At
CesA1 may be required (Desprez
et al., 2002).
Brittle culm mutants have been identified in barley, maize, and rice. The
cellulose content in the cell walls of cells in the brittle culm mutants of barley
was found to be lower than the wild-type plants, but no significant differences
were found in the amount of the noncellulosic components of the cell wall
(Kokubo
et al., 1989, 1991). Brittle culm mutants in rice were useful in identifying
three
CesA genes (Os
CesA4, Os
CesA7, and Os
CesA9) (Tanaka
et al., 2003). The three
genes are expressed in seedlings, culms, premature panicles, and roots, but not in
mature leaves. The expression profiles are almost identical for these three genes,
and decrease in the cellulose content in the culms of null mutants of the three
genes indicates that these genes are not functionally redundant (Tanaka
et al.,
2003).
Identification of Other Genes/Proteins Which May be Required for
Cellulose Biosynthesis in Plants
The role of β-1,4-endoglucanase during cellulose synthesis was first proposed by
Matthysse
et al. (1995a,b) during analysis of cellulose-minus mutants in
Agrobacterium tumefaciens (Matthysse
et al., 1995a,b). In this bacterium, cellulose synthesis
is suggested to proceed via the formation of lipid-linked intermediates, and a
β-1,4-endoglucanase is predicted to function as a transferase in the transfer of
β-1,4-linked glucan oligomers from a lipid carrier to the growing cellulose chain
(Matthysse
et al., 1995a). The gene encoding β-1,4-endoglucanase is organized
with the cellulose synthase gene in an operon in A.
tumefaciens, and a similar
organization of these genes is observed in a number of other bacteria (Matthysse
et al., 1995b; Römling, 2002). The organization of a β-1,4-endoglucanase gene with
the cellulose synthase gene in the same operon in bacteria has been taken as an
indication that β-1,4-endoglucanase probably has a role during cellulose synthesis.
So far, there is no direct demonstration for this role in bacteria or any other
organism. A gene encoding a membrane-anchored β-1,4-endoglucanase called
Korrigan also has been identified in a dwarf mutant of
Arabidopsis (Nicol
et al., 1998). In plants, the
Korrigan protein is believed to
function during primary or secondary wall cellulose synthesis (Lane
et al., 2001;Mølhøj
et al., 2002; Nicol
et al., 1998; Sato
et al., 2001; Szyjanowicz
et al., 2004; Zuo
et al., 2000). Its exact function during cellulose synthesis remains to be determined,
although various roles have been assigned to it such as terminating or editing the
glucan chains emerging from the cellulose synthase complex before their crystallization
into a cellulose microfibril. Alternately it could cleave sterol from the
sterol-glucoside primer that is suggested to initiate glucan chain formation (Peng
et al., 2002). However, recent evidence does not support this role (Scheible and
Pauly, 2004). A membrane-bound sucrose synthase, which converts sucrose to
UDP-glucose, may be physically linked to the cellulose synthase complex
for channeling UDP-glucose to the cellulose synthase in plants, and suppression
of this gene has been shown to effect cotton fiber initiation and elongation
(Amor
et al., 1995; Ruan
et al., 2003).
Proteins that may indirectly influence cellulose biosynthesis include those that
are required for N-glycan synthesis and processing (Lukowitz
et al., 2001). One of
these proteins is glucosidase I, which trims off the terminal β-1,2-linked glucosyl
residue from N-linked glycans and is involved in the quality control of newly
synthesized proteins that transit through the endoplasmic reticulum (ER)
(Boisson
et al., 2001; Gillmor
et al., 2002). Another protein could be glucosidase II
that removes the two internal β-1,3-linked glucosyl residues subsequent to the
action of glucosidase I in the quality control pathway (Burn
et al., 2002b). Other
proteins that influence cellulose production include KOBITO, a membraneanchored
protein of unknown function that is suggested to be a part of the
cellulose synthase complex, and COBRA, a putative glycosylphosphatidylinositol
(GPI)-anchored protein, which upon being inactivated, dramatically reduces culm
strength in rice (Li
et al., 2003b; Pagant
et al., 2002; Schindelman
et al., 2001).





