Techniques to Distinguish Nonfermentative Gram-Negative Bacilli from Enterobacteriaceae
A variety of gram-negative bacilli that normally inhabit soil and water or live as commensals on human mucous membranes may contaminate specimens sent to the microbiology laboratory for culture or, more importantly, may produce opportunistic human infections. Although the Gram-stain appearance and cultural characteristics of the organisms may resemble those of Enterobacteriaceae, they are relatively inactive in the common biochemical tests. In particular, they either fail to metabolize glucose or they degrade it by oxidative rather than fermentative pathways. For this reason these organisms are often referred to as “glucose nonfermenters” (as opposed to the glucose-fermenting enteric bacilli). A number of bacterial genera and species are included in this group of nonfermenters. The most important from a medical aspect is Pseudomonas aeruginosa, which is most often involved in human infection. Because of the different clinical implications and the varying antimicrobial susceptibility patterns (nonfermenters are more highly resistant to common antimicrobial agents) it is important to distinguish nonfermenters from enteric bacilli. The characteristics of a few nonfermenting bacteria are listed in table 24.5-1 and compared with those of the Enterobacteriaceae.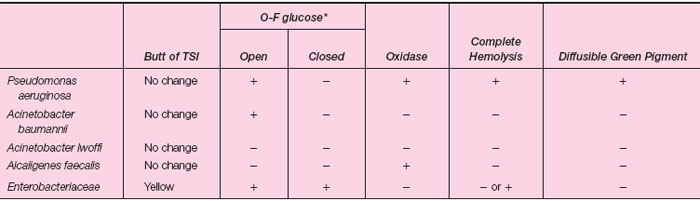 |
| Table 24.5-1 Characteristics of Nonfermenting Gram-Negative Bacilli |
*A positive test is a yellow color. Yellow in the open tube only indicates glucose degradation or oxidation. A yellow color in the closed tube (with mineral oil) indicates the organism is fermentative rather than oxidative. Glucose fermenters produce acid (yellow color) in the open as well as the closed tube.
| Purpose | To study some biochemical reactions of glucose nonfermenting bacteria |
| Materials | Blood agar plates Nutrient agar plates TSI slant O-F glucose deeps Oxidase reagent (di- or tetramethyl-p-phenylenediamine) Dropper bottle with sterile mineral oil Slant cultures of Pseudomonas aeruginosa,Acinetobacter baumannii, and Escherichia coli |
Procedures
- Prepare and examine a Gram-stained smear of each organism.
- Inoculate a blood and nutrient agar plate with each organism. Streak the plate to obtain isolated colonies.
- Inoculate each organism onto a TSI slant by stabbing the butt and streaking the slant.
- Inoculate two tubes of O-F glucose with each organism by stabbing your inoculating loop to the bottom of the column of medium. Overlay one of each set of two tubes with a one-half inch layer of sterile mineral oil.
- Label all plates and tubes. Incubate them at 35°C for 24 hours.
- Test each organism for the presence of the enzyme oxidase. The procedure is as follows.
- Take a sterile petri dish containing a piece of filter paper.
- Wet the paper with oxidase reagent.
- With your inoculating loop, scrape up some growth from the tube labeled P. aeruginosa and rub it on a small area of the wet filter paper. You should see an immediate positive oxidase reaction as the color of the area changes from light pink to black-purple.
- Repeat procedure 6c using growth from the tubes labeled A. baumannii and E. coli. Record the results in the table.
Results
- Examine the blood agar plate for hemolysis and the nutrient agar for pigment production.
- Read and record all biochemical reactions in the following table.
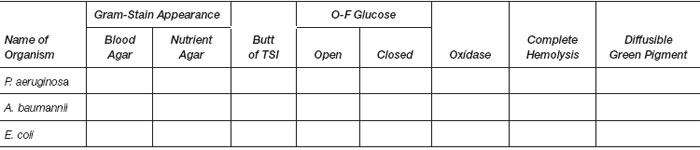 |




