Glutamine, Glutamate, Aspartate, and Asparagine are Central Regulators of Nitrogen Assimilation, Metabolism, and Transport
Glutamine, glutamate, aspartate, and asparagine constitute a metabolic network
[hereafter termed for simplicity ‘‘amide amino acid metabolism’’ (AAAM)
because it contains the two amide amino acids, glutamine and asparagine] that
participates in numerous processes (Fig. 3.1). These include nitrogen assimilation,
nitrogen metabolism into the various amino acids and other nitrogenous compounds,
nitrogen transport between sources and sinks, carbon/nitrogen partitioning,
and stress-associated metabolism. The AAAM network is regulated in a
concerted manner by numerous metabolites and environmental signals, such as
by light and phytochrome, in a manner that varies significantly between different
plant tissues and organs, as well as in response to developmental, physiological,
and environmental signals. Ammonium ion, derived either from nitrogen assimilation
or from photorespiration, is incorporated into glutamine by a reaction
catalyzed by glutamine synthase (GS), and glutamine is further converted into
glutamate catalyzed by glutamate synthase (GOGAT) (Fig. 3.1). Glutamate is
trans-aminated to aspartate by a large family of aspartate amino transferases
and aspartate can be converted into asparagine and back from asparagine into
aspartate by the activities of asparagine synthetase and asparaginase, respectively
(Fig. 3.1). Glutamine, glutamate, and aspartate are used for the synthesis of other protein and nonprotein amino acids, as well as amides and other nitrogenous
compounds. Asparagine, which is synthesized from aspartate, serves not only as
a protein amino acid but is also as a major nitrogen transport agent. The regulation
of nitrogen assimilation and metabolism in plants has been discussed in detail in a number of reviews (Hirel and Lea, 2001; Ireland and Lea, 1999; Lam
et al.,
1995; Lea and Ireland, 1999; Miflin and Habash, 2002; Oliveira
et al., 2001;
Stitt
et al., 2002).
In this chapter, we focus mainly on studies dealing with genetic engineering of
enzymes associated with AAAMand analysis of plant mutants. However, several
principles of AAAM are important for understanding its functional significance
and the enzymes that control this metabolic network (Stephanopoulos, 1999).
In this context, the synthesis of amino acids requires both carbon and nitrogen
and is therefore regulated in a concerted manner by nitrogen and sugars (Singh,
1999). When nitrogen and sugar levels are not limiting, the assimilated nitrogen
triggers sugar metabolism to efficiently synthesize glutamine and glutamate and
the synthesis of other amino acids. However, when carbon levels are limiting
(termed carbon starvation), glutamine and glutamate are efficiently converted
into sugars, while the released nitrogen is stored in nitrogen-rich metabolites,
such as asparagine and arginine (Coruzzi and Last, 2000). In nonsenescing tissues,
amino acid metabolism is subject to a tight diurnal regulation. During daytime,
when photosynthesis is active, glutamine, glutamate, and aspartate are
used efficiently for synthesis of other amino acids needed for protein synthesis,
while during the night these amino acids are strongly converted into asparagine
serving as a nitrogen storage and transport compounds (Morot-Gaudry
et al., 2001).
In senescing tissues,
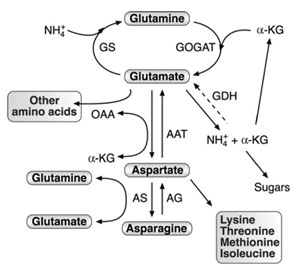 |
| FIGURE 3.1 Schematic diagram of the network
of AAAM and its connection to nitrogen
assimilation,
carbon metabolism, and synthesis of
other amino acids. Abbreviations: GS, glutamine
synthetase; GOGAT, glutamate synthase; AAT,
aspartate amino transferase; GDH, glutamate
dehydrogenase; AS, asparagine synthetase; AG,
asparaginase; OAA, oxaloacetate; α-KG,
α-ketoglutarate. The dashed arrow represents the
aminating activity of GDH, which was
experimentally demonstrated in plants, but its
function is still a matter of debate. |
the AAAMnetwork is used to convert the various amino acids
and ammonium ion, which are derived from protein breakdown (particularly
RuBisCO and other major plastid-localized photosynthetic genes), into transport competent nitrogenous compounds, such as asparagine, glutamine, and ureides
(Hirel and Lea, 2001; Ireland and Lea, 1999; Lam
et al., 1995; Lea and Ireland, 1999;
Miflin and Habash, 2002). These processes take place by the activation of many
amino acid catabolism pathways as well as enzymes of AAAM. Under stress
conditions, the AAAM network is used for rapid production of stress-associated
metabolites, such as proline, arginine, polyamines, and γ-amino butyric acid.
Hence, AAAMis a most highly controlled metabolic networks in plants.