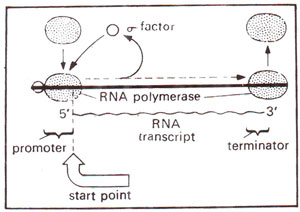
Fig. 32.5. A transcription unit, showing promoter region, start point and terminator; also shown is the role of RNA polymerase with its core enzyme and sigma factor.
The elongation of the transcript may be impeded by (i) 'pause sites', (ii) 'arrest sites' or 'dead ends' and (iii) 'terminators' or 'release sites' (Fig. 32.5). The
'pause sites' induce a temporary reversible block to nucleotide addition. The
'arrest sites' stop elongation, to be resumed only in the presence of elongation factors
GreA and
GreB (in eukaryotes the corresponding transcription factor is TFIIS described later in this section), which assist in a novel transcript cleavage reaction (Fig. 32.27). The
'release sites' lead to release of RNA and/or
RNAP either intrinsically, or upon activation by factors like NusA,
Rho, or
Tau.

Fig. 32.5. A transcription unit, showing promoter region, start point and terminator; also shown is the role of RNA polymerase with its core enzyme and sigma factor.
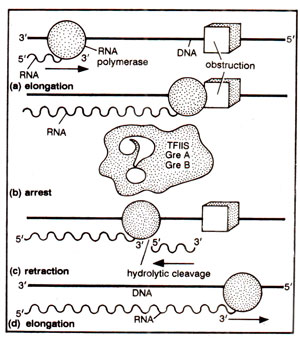
Fig. 32.27. The role of TFIIS in elongation of RNA chain

Fig. 32.27. The role of TFIIS in elongation of RNA chain
Although the nature of DNA sequences causing transcription arrest is not known, the mechanism of transcription arrest and the roles of GreA and GreB in restoring elongation can be explained in the framework of '
inchworm model' described above. The transcription arrest may result when the catalytic site of RNAP is misplaced from the 3' end of growing transcript. This, itself may result, if the '
lagging product site' slipped backward on the template, bringing the catalytic site out of register. Transcript cleavage by GreB would create a new 3' end into the right register, thus restoring elongation. Further, at some template sites, translocation may disturb the placement of catalytic site into the right register, and GreA-induced cleavage of short segments might restore the appropriate register to resume elongation. Thus an elongation barrier may be overcome by RNAP through a 'backup and restart' mechanism.







