Chi-square test
In the beginning of this section, we described two examples of linkage, one each in sweet pea and maize, where results of a dihybrid test cross (AaBb x aabb) deviated from 1AB : 1Ab : 1aB : 1ab ratio expected due to independent assortment. In experiments like these, when there are deviations from expected 1:1:1:1 ratio or 9 : 3 : 3 : 1 ratio, it may not be easy to decide on the presence or absence of linkage by simply looking at the data. Instead an analytical approach may be needed. The chi-square (χ2) test is one such approach, which is used firstly, for testing the goodness of fit to an expected ratio and secondly, for the detection of linkage in more certain terms. This test tells us how often deviations like those being examined will occur purely on the basis of chance.
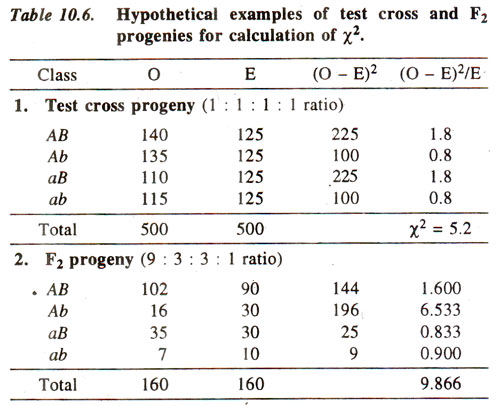
(ii) Calculate χ2. This value is calculated from observed (O) results, using actual numbers (not from percentages or fractions) and expected (E) results calculated from total population using 1:1:1:1 ratio or 9 : 3 : 3 : 1 ratio (whichever is the case). Following formula is used :
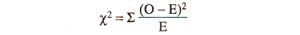
(where σ refers to sum of values of χ2, over all classes) The use of this formula for test cross progeny (1:1:1:1 ratio) and F2 progeny (9:3:3:1 ratio) is illustrated in Table 10.6.
(iii) Work out degrees of freedom. The degrees of freedom (df) can be worked out as follows :
df = (number of classes - 1)
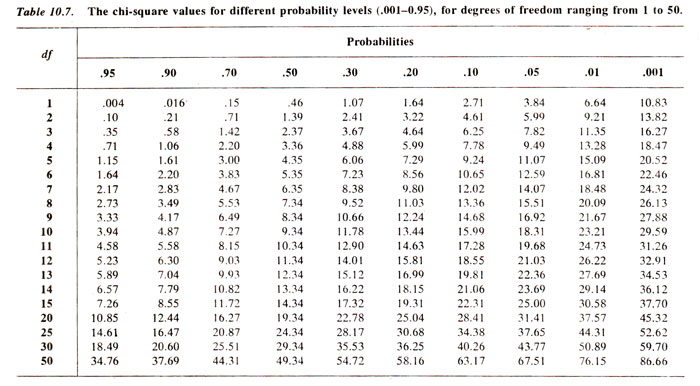
(v) Reject or accept the null hypothesis. We generally use an arbitrary value of p = 0.05 = 5%, so that if the computed value of χ2. is below 5% level of probability, we reject the null hypothesis, otherwise accept it. In the examples as above, for test cross progeny, χ2 = 5-2 has a p > 10%. Therefore, null hypothesis is accepted, suggesting that the ratio 1:1:1:1 holds good. On the other hand, for F2 progeny, χ2 = 9.866 has a p < 5% suggesting that the ratio 9:3:3:1 does not hold good.
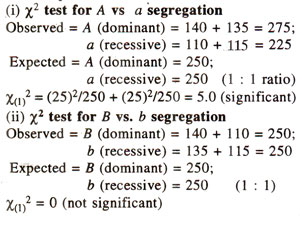
Partitioning of χ2 value for a dihybrid test cross progeny. The χ2 value of 5.2 for 3 degree of freedom obtained in Table 10.6 for 1 : 1 : 1 : 1 ratio can be partitioned as follows :
For this test, expected ratios are not available, but expected values for each class can be calculated, assuming that segregation at one locus (A vs. a) is not dependent (contingent) upon the segregation at the second locus (B vs. b). The test is called 'test for independence or contingency test'. For conducting this test, the data from Table 10.6, is rearranged in a contingency table as follows :
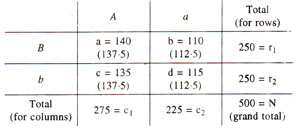
where,
a = AB r1 = first row total
b = aB r2 - second row total
c = Ab c1 = first column total
d = ab c2 = second column total
degrees of freedom for χ2 = (r - 1) (c - 1) = 1
(r = number of rows; c = number of columns)
In the above contingency table, the expected values (a^,b^,c^,d^) of a, b, c, and d are given in parentheses and can be calculated as follows :
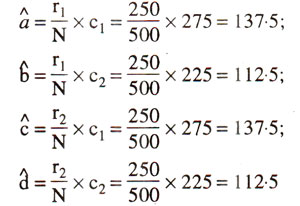
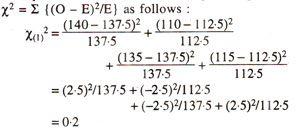
The chi-square for independence can also be calculated using the following equation, so that expected values need not be calculated.
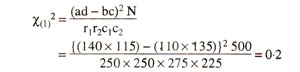
However it is recommended that, when any one of the four observed values (a, b, c, d) is below 5, a Yate's correction for continuity should be applied, and the equation is modified as follows :
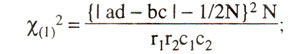
The above partitioning of χ2 demonstrates clearly that although there is a good fit to the ratio 1:1:1:1, but there is some distortion in segregation of A vs. a, and that there is no evidence of linkage.
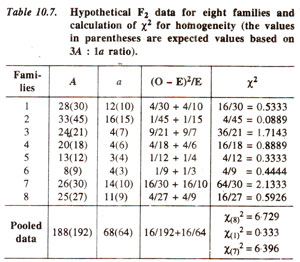
Partitioning of χ2 value for a dihybrid F2 population. The chi-square value for goodness of fit to 9 : 3 : 3 : 1 ratio expected in F2 population derived due to selfing of a F1 dihybrid (AaBb) can also be partitioned. As an illustration, the chi-square value of 9.866 for three degrees of freedom calculated for 9:3:3:1 ratio in Table 10.6 is significant at 5% level. This can be partitioned as follows :