Chloroplast genetics Non-chromosomal genes in Chlamydomonas

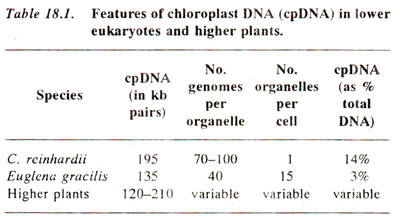
Fig. 18.16. Uniparental inheritance in a haploid organism like Chlamydomonas, although cytoplasm is transmitted from both the gametes.
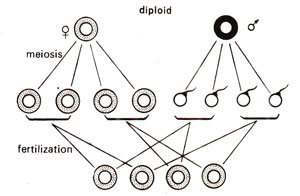
Fig. 18.17. Maternal inheritance in a diploid organism, where cytoplasm is transmitted only through the egg.
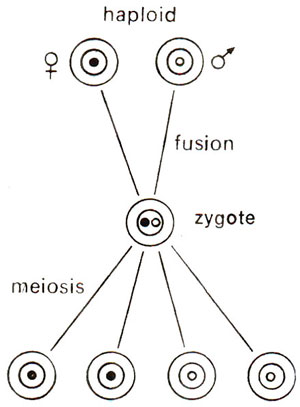
Fig. 18.18. Segregation of nuclear genes in a haploid organism (1 : 1 ratio).
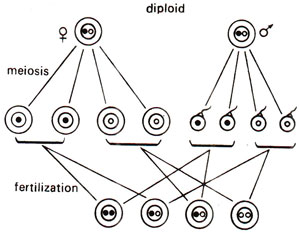
Fig. 18.19. Segregation of nuclear genes in a diploid organism (1:2:1 ratio).

Fig. 18.20. CsCl density gradient centrifugation of Chlamydomonas DNA, showing cpDNA band distinct from nuclear DNA band.
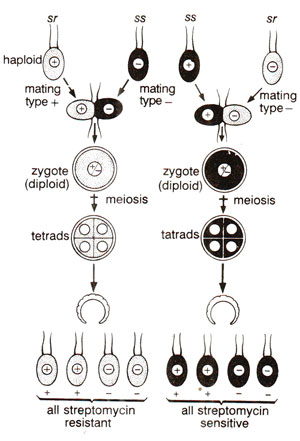
Fig. 18.21. Reciprocal cross (sr mt+ x si mt- and ss mt+ x sr mt-), showing uniparental inheritance of non-chromosomal genes and lack of segregation (sr = streptomycin resistance and ss = sensitivity to streptomycin) along with segregation of nuclear genes (mt+ and mt-).
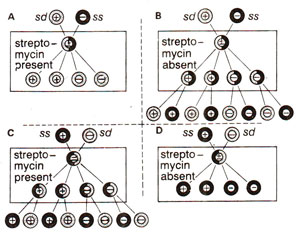
Fig. 18.22. Results of reciprocal crosses (sd mt+ x ss mt- ; ss mt+ x sd mt-), when the progeny is grown under two conditions (streptomycin present or streptomycin absent). Unfavourable conditions (absence of streptomycin in B and presence of streptomycin in C) lead to selection of exceptional biparental zygotes (for details see text).
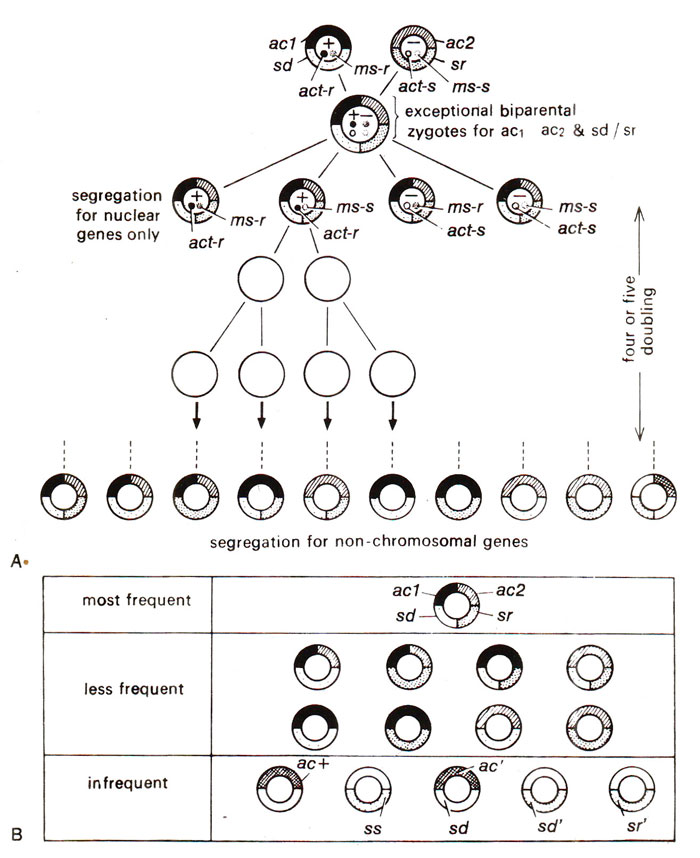
Fig. 18.23. A cross involving two non-chromosomal (ac1/ac2 and sd/sr) and three nuclear genes (act-r/act-s; ms-r/ms-s; mt+/mt-) showing segregation of nuclear genes soon after meiosis and that of non-chromosomal genes in the following generations (A); relative frequencies of different cytoplasmic types obtained in this cross (B); act+, ac', ss, sd', sr' represent new types (results of only 1% exceptional biparental zygotes are shown—for details see text).
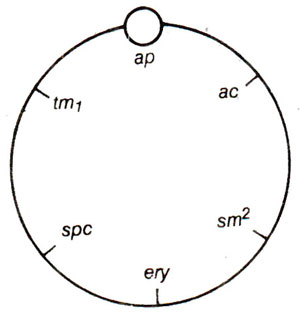
Fig. 18.24. A circular genetic map of chloroplast genome in Chlamydomonas.

Fig. 18.25. Map of circular chloroplast DNA of Zea mays, showing the positions of duplicate inverted region of rRNA genes and also RuBP (ribuIose bisphosphate) carboxylase gene (after Bedbrook et at, 1979).
(a) Uniparental inheritance.
As shown earlier, several characters show maternal inheritance both in plants and animals. This was earlier explained to be due to greater amount of cytoplasm in egg than in sperm or male gamete. In Chlamydomonas, which is a haploid organism, sexual reproduction is isogamous, both gametes being similar in morphology and thus having equal amounts of cytoplasm. Despite this lack of difference in cytoplasmic content, in Chlamydomonas, some characters are transferred only from one parent, exhibiting what is called uniparental inheritance, rather than maternal inheritance, because in Chlamydomonas, we describe two sexes as two mating types, i.e., mt+ (plus) and mt- (minus), rather than as female and male (Fig. 18.16). One may notice that in diploids, the male gametes do not contribute cytoplasm (Fig. 18.17). One may also distinguish between the patterns of inheritance between non-chromosomal and chromosomal genes. The inheritance patterns for chromosomal genes in haploid and diploid organisms are shown in Figures 18.18 and 18.19.
Fig. 18.16. Uniparental inheritance in a haploid organism like Chlamydomonas, although cytoplasm is transmitted from both the gametes.

Fig. 18.17. Maternal inheritance in a diploid organism, where cytoplasm is transmitted only through the egg.

Fig. 18.18. Segregation of nuclear genes in a haploid organism (1 : 1 ratio).

Fig. 18.19. Segregation of nuclear genes in a diploid organism (1:2:1 ratio).
Uniparental inheritance has also been demonstrated at the level of cpDNA, due to the following facilities : (i) cpDNA forms a separate distinct band on CsCl density gradient centrifugation (Fig. 18.20); (ii) the cpDNA of two parents can be differently labelled with N14 and N15, the two giving buoyant densities of 1.69 and 1.70 respectively; this difference looks small, but they form separate bands on CsCl density gradient. It was shown that the buoyant density of zygote always resembled that of mt+ (female) parent and that cpDNA of mt- parent is lost, inactivated or destroyed. Restriction enzyme patterns of cpDNA in zygote also resembled those of mt+ parent suggesting uniparental transmission of cpDNA.

Fig. 18.20. CsCl density gradient centrifugation of Chlamydomonas DNA, showing cpDNA band distinct from nuclear DNA band.
(b) Segregation, independent assortment and gene mapping.
In earlier years (before 1960), a large number of cases were known, where non-chromosomal inheritance, as evident from uniparental or maternal inheritance, were described. However, further detailed study of these traits was not possible. There were two reasons for this. First, the mutant non-chromosomal genes could not be artificially produced when desired and spontaneous mutations for these genes were rare, thus making their study difficult. Second, since these traits were transmitted from only one parent, standard techniques of genetic analysis could not be applied to study segregation and independent assortment as studied in chromosomal genes. Ruth Sager was successful in overcoming both these difficulties in Chlamydomonas and was thus able to study the non-chromosomal genes in this organism in some detail.Ruth Sager found that if streptomycin sensitive cells are grown in a medium containing streptomycin, several kinds of induced mutations in chloroplast including streptomycin resistant mutations were produced. Streptomycin resistance could be found on chromosomes also where it may be inherited in a Mendelian manner, but in some strains this character showed uniparental inheritance as shown in Figure 18.21. It may be noticed that when the mt+ strain was streptomycin resistant, all the progeny was streptomycin resistant and when it was sensitive, all the progeny sensitive. Streptomycin was used for inducing not only resistance to streptomycin but also to induce mutations for several other non-chromosomal traits thus making it possible to study their inheritance.

Fig. 18.21. Reciprocal cross (sr mt+ x si mt- and ss mt+ x sr mt-), showing uniparental inheritance of non-chromosomal genes and lack of segregation (sr = streptomycin resistance and ss = sensitivity to streptomycin) along with segregation of nuclear genes (mt+ and mt-).
The second difficulty of getting the transmission of non-chromosomal genes from only one parent was overcome when it was observed by Ruth Sager that in very rare cases streptomycin resistance and streptomycin sensitivity from parents of both mating types, mt+ and mt- could be transmitted. It was found that, when streptomycin dependent strain (sd, which will not survive without streptomycin) was used as mt+ strain, and progeny grown in a medium containing streptomycin, only sd strains will be produced (Fig. 18.22 A). However, if the same progeny is grown in a medium lacking streptomycin, then biparental exceptional zygotes will grow, because the uniparental sd strains will not survive in the absence of streptomycin (Fig. 18.22 B). Similarly in the reciprocal cross, using ss (streptomycin sensitive) as the mating type mt+ and sd as mt-, only ss strains will be available in the absence of streptomycin showing uniparental inheritance (Fig. 18.22 D), but, if the progeny is grown in the presence of streptomycin, then biparental zygotes will be produced (Fig. 18.22 C). Later it was discovered that if mt+ parent is irradiated with ultraviolet (UV) irradiation prior to mating, frequency of biparental zygotes will greatly increase. In this manner, techniques for obtaining exceptional biparental zygotes could be standardized.

Fig. 18.22. Results of reciprocal crosses (sd mt+ x ss mt- ; ss mt+ x sd mt-), when the progeny is grown under two conditions (streptomycin present or streptomycin absent). Unfavourable conditions (absence of streptomycin in B and presence of streptomycin in C) lead to selection of exceptional biparental zygotes (for details see text).
As shown above, Ruth Sager could overcome the two difficulties for genetic analysis of chloroplast genes. After having achieved this, Sageri and her co-workers made crosses between strains carrying contrasting characters controlled by non-chromosomal genes. This will be illustrated with the help of an example used by her in her article published in Scientific American in 1965. The mt+ parent carried ac1 (acetate requiring mutant) and sd (streptomycin dependence), and mtr parent carried ac2 and sr (the other alleles of same genes ac1 and sd).
It was found that 99 per cent progeny though segregated for nuclear genes act-r/act-s, ms-r/ms-s and mt+/mt- but they all showed maternal or uniparental transmission of ac1 and sd. One per cent exceptional biparental zygotes were obtained, which were selected and their behaviour in future generations was studied to explain the pattern of inheritance of non-chromosomal genes. The details of the results are shown in Figure 18.23. It was shown that during meiosis the nuclear genes segregated, but non-chromosomal genes did not segregate immediately, and segregated slowly in the following generations. The following conclusions were made on the basis of the results shown in Figure 18.23.

Fig. 18.23. A cross involving two non-chromosomal (ac1/ac2 and sd/sr) and three nuclear genes (act-r/act-s; ms-r/ms-s; mt+/mt-) showing segregation of nuclear genes soon after meiosis and that of non-chromosomal genes in the following generations (A); relative frequencies of different cytoplasmic types obtained in this cross (B); act+, ac', ss, sd', sr' represent new types (results of only 1% exceptional biparental zygotes are shown—for details see text).
- The non-chromosomal or chloroplast genes segregate like nuclear genes in 1 : 1 ratio in mitotic divisions, so that ac, and ac2 as well as sd and sr behaved as alleles.
- The ac1/ac2 and sd/sr gene pairs segregate independently of each other, in both time and space, giving rise to parental combinations ac1 sd and ac2 sr as well as the recombinants ac1 sr and ac2 sd.
- Even wild type or new cytoplasmic alleles arose, which are designated as ac+, ac', ss, sd' and sr'. These new alleles are believed to have originated due to intragenic recombination discussed in Fine Structure of Gene-at the Genetic Level (A New Concept of Allelomorphism) for nuclear genes.
A circular genetic map of chloroplast genome of Chlamydomonas is shown in Figure 18.24, where an attachment point ap is also shown. (For details consult Ruth Sager's book).
Chloroplast genetics in higher plants.
We know that chloroplasts of lower eukaryotes and higher plants like Zea mays contain DNA. Chloroplast genomes are relatively large (with a narrow range) usually ranging from 120 kb to 210 kb (kb = kilo base pairs) except in Acetabularia (400 kb). This is comparable to the size of a large bacteriophage like T4 (110 kb). Each cell may have several chloroplasts and each chloroplast may have several circular DNA molecules, so that the chloroplast DNA (cpDNA) may make upto 14% of cellular DNA in lower eukaryotes. In higher eukaryotes, however, the cpDNA makes a small proportion of total DNA in the cell. These features of chloroplast DNA are presented in Table 18.1.
Fig. 18.25. Map of circular chloroplast DNA of Zea mays, showing the positions of duplicate inverted region of rRNA genes and also RuBP (ribuIose bisphosphate) carboxylase gene (after Bedbrook et at, 1979).
Chloroplast genome and prokaryotes (a comparison).
A detailed study of chloroplast genome in eukaryotes, has revealed that the chloroplast genomes share both prokaryotic and eukaryotic features. Therefore, the view that they are the remnants of entrapped prokaryotic genomes is an oversimplification. Some of the features in which chloroplasts resemble the prokaryotes and those in which they differ are as follows—(for more details consult Expression of Gene : Protein Synthesis 4. Translation in Prokaryotes and Eukaryotes).(a) Resemblances between chloroplast and prokaryotes.
- The ribosomes are 70S in size in both the cases.
- The rRNA genes as well as several tRNA genes have very high degree of homology as revealed by sequence homology.
- Preceding the initiation codon AUG in chloroplasts, there are present ribosome binding sites, and the sequences which bear strong resemblance to 'Pribnow Box' (-10 and the -35 regions) of bacteria (consult Expression of Gene : Protein Synthesis 2. Transcription in Prokaryotes and Eukaryotes and Regulation of Gene Expression 1. Operon Circuits in Bacteria and other Prokaryotes).
- There is sequence overlapping between some genes in chloroplasts, a feature known in prokaryotes. In chloroplasts it can not be due to economy since non coding sequences are also available in chloroplasts.
- There is a resemblance between RNA polymerases of chloroplasts and prokaryotes. For instance, in Chlamydomonas reinhardii the enzyme is sensitive to rifampicin and contains a a (sigma) like polypeptide. There is also evidence of homology between p and p' subunits of E. coli RNA polymerase and those of C. reinhardii chloroplast RNA polymerase.
- Protein synthesis in chloroplasts can be inhibited by chloramphenicol as in bacteria, since only 70S ribosomes are sensitive to this drug. Cycloheximide does not inhibit protein synthesis in chloroplasts, as it does in the cytoplasm (SOS ribosomes) of eukaryotes.
- Initiation of protein synthesis takes place by formyl methionyl tRNA both in chloroplasts and prokaryotes.
(b) Differences between chloroplast and prokaryotic genomes.
- In chloroplasts there are split genes, e.g. (i) rRNA genes and protein genes in chloroplasts of C. reinhardii, (ii) protein genes in chloroplasts of Euglena and (iii) tRNA genes of several chloroplasts of higher organisms. The introns of these split genes fall in three groups and differ from those in nuclear genes of eukaryotes (consult Organization of Genetic Material 3. Split Genes, Overlapping Genes and Pseudogenes).
- Chloroplast DNA sequences have the ability to promote autonomous replication. This feature is absent in prokaryotes.
- Presence of short repetitive elements interspersed throughout several, but not all chloroplast genomes is also a eukaryotic feature.