Donor (F+ or Hfr) and recipient (F-) strains (discovery of fertility factor or sex factor F)
Among the above strain A
(met-bio-thr+leu+thi+) and strain B
(met+bio+thr-leu-thi-), undergoing recombination, it was possible to designate them as donor and recipient strains on the basis of the following simple experiments involving treatment of either A or B with streptomycin (which does not kill immediately, but prevents cell division). When strain A was treated with streptomycin and mixed with strain B after washing out streptomycin, the frequency of colonies on minimal medium equaled those in control, where both strains were untreated. On the other hand,
when only strain B was similarly treated, no colonies were observed, suggesting that for recombination, cell division in strain B, but not in strain A, was essential. This suggested that gene transfer in
E. coli is non-reciprocal and that one strain (A) is donor and the other (B) is recipient, which has to undergo cell division to form colonies after receiving genes from the donor strain (A). This differs from sexual reproduction where two parents contribute genetic material to an entirely new organism, the parents themselves remaining unaltered. Later on, through a series of experiments, it could be proved that the donor attribute of a strain is due to a transmissible fertility factor, F, which is absent in the recipient strain. Therefore, the donor was designated as F
+ and recipient s F
-. It was also shown that a F
+ strain can also act as a recipient, although F
- can not act as a donor unless it changes to F
+ through conjugation with a F
+ strain.
The F factor is composed of a circular DNA molecule of molecular weight 250,000 (about l/40th of bacterial chromosome). Ordinarily only one F factor is found per cell (Fig. 12.5), which replicates with each cell division. A donor cell or F
+ can transmit F factor to recipient or F
- cells which in turn become donor or F
+ cells. This transmission of F factor is independent of transmission of chromosomal genes, so that in many cases F factor may be transmitted in high frequency while chromosomal genes are transmitted in a very low frequency (one per million cells). However, rarely from F
+ populations, specific donor strains capable of transmitting chromosomal genes in high frequency (a thousand times that of F
+), could be isolated.
These were isolated for the first time by
Cavalli-Sfroza and were called high frequency recombinants and designated as
Hfr strains. Besides, higher frequency of recombination in Hfr, there is another important difference between F
+ and Hfr strains. The initial transfer of genes from a Hfr to a F
- strain was not random but the genes were transferred in a regular and definite order in the form of a linkage group. To illustrate this further, let us suppose that there are chromosomal genes
A to
Z on bacterial chromosome. The F
+ strains could produce recombinants for all genes in approximately equally low frequency. On the other hand a particular Hfr strain,
transmits different genes in different frequencies and for a particular Hfr strain, the order of genes showing high to low recombination values is fixed. However, the order of genes showing high to low recombination values differs in different Hfr strains so that while in one Hfr strain,
A may show the highest recombination value, in other strains
K or
P or
Z may show highest recombination (Fig. 12.6). But once the gene showing highest frequency is known, the order in which other genes will be transferred, can be easily predicted as shown in Figure 12.7. This aspect is illustrated in Figure 12.8, using a specific Hfr strain, where the genes are transferred in the order
azir > tanr > lac+ > gal+. Moreover, while most of the F
- conjugating with F
+, become F
+, only very rarely F
- conjugating with Hfr would become F
+ or Hfr. This difference in the behavior or F
+ and Hfr has been explained in the following manner.
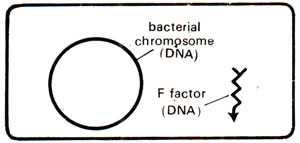
Fig. 12.5. A bacterial cell showing circular DNA and F factor.
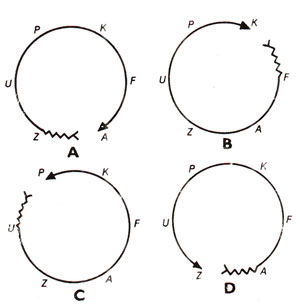
Fig. 12.6. (A, B, C, D). Four hypothetical Hfr strains having different genes at their proximal ends.

Fig. 12.7. The linear order of genes in five hypothetical Hfr strains, resulting due to breakage of bacterial chromosome at different sites and subsequent orientation acquired by the resulting linear chromosome
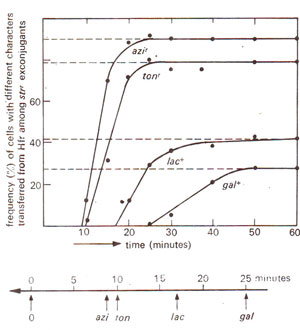
Fig. 12.8. (a) Results of a cross sirr, azis, tons, lac-, gal-(F ) x strs, azir, tonr, lac+, gal+ (Hfr), where samples from incubated mixture were drawn at different time intervals, disrupted in a blender, and spread on a medium containing streptomycin. The resistant colonies (sirr) were derived from F- parent (due to transfer of genes from Hfr) and were tested for four metabolic steps, (b) Chromosome map prepared from results shown in (a).

Fig. 12.9. Interconvefsion of F+ and Hfr strains, due to integration and detachment of F factor.

Fig. 12.10. Sexual conjugation, showing two alternatives. In one case (right) the F factor remains free, while in the other case (left) the F factor gets attached to form Hfr.
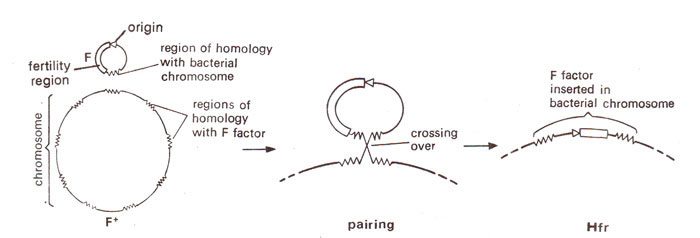
Fig. 12.11. A model for insertion of F factor into E. coli chromosome; an area of fertility factor has several regions of homology in E. coli chromosome, permitting crossing over (regions of homology are represented by jagged lines).

Fig. 12.12. Origin of F' factor (or F-lac) due to abnormal oullooping (a-c) and reintegration of F-lac+ into F-lac-strain.
The F factor remains free in the cytoplasm in F
+ strains but remains integrated with the chromosome in Hfr strains. The position at which the F factor will be attached to chromosome differs in different Hfr strains. Also F
+ can become Hfr due to integration of F factor to the chromosome and vice versa (Fig. 12.9).

Fig. 12.9. Interconvefsion of F+ and Hfr strains, due to integration and detachment of F factor.
When conjugation occurs between F
+ donor and F
- recipient, two possibilities exist (Fig. 12.10) (i) F factor may remain free in cytoplasm, so that only F factor will enter the recipient cell converting the recipient F
- into a donor F
+. (ii) F factor may get integrated and would thus make the distal part of bacterial chromosome. In this case the chromosome will become linear and would have a directional orientation, so that it would enter the recipient cell from the end away from attached F factor.

Fig. 12.10. Sexual conjugation, showing two alternatives. In one case (right) the F factor remains free, while in the other case (left) the F factor gets attached to form Hfr.
From the above discussion, it also emerges that the low recombination attribute of F
+ strain is due to presence of Hfr among F
+ cells in low frequency. When same Hfr is found in a relatively pure state, recombination frequency increases. In other words, for recombination to take place, F factor must be attached to bacterial chromosome and the circular chromosome should take up a linear orientation.
Conversion of F+ into Hfr strain
It has been shown that like the bacterial chromosome, fertility factor F is also circular, so that a crossover between the two rings would produce a larger ring with F inserted. The F is believed to consist of three regions : (i) origin, (ii) fertility genes and (iii) pairing region. The pairing region of F has homology with several regions of bacterial chromosome, so thatcrossing over can take place at any one of these regions, producing different Hfr strains (Fig. 12.11).

Fig. 12.11. A model for insertion of F factor into E. coli chromosome; an area of fertility factor has several regions of homology in E. coli chromosome, permitting crossing over (regions of homology are represented by jagged lines).
Fertility factor F' and sexduction
During experiments with Hfr strains, initially (in 1959) an Hfr strain was found, which kept on producing F
+ cells at high frequencies. The fertility factor involved in this process was described as F' to distinguish it from normal F,
due to following attributes : (i) F'-bearing F
+ strains often reverted back to an Hfr strain; (ii) F' always integrated at the same location to give the same original Hfr (not the different Hfr strains having different proximal and distal genes, as is usually observed). The origin of F' was resolved through the following interesting observations. From a Hfr
lac+ (with
lac+ at distal end), an F
+ strain was derived, which transferred
lac+ to
F-lac-at a high frequency and
F-lac- became
F+lac+, which was shown to be
(F-lac+)/lac-(
lac+carried F, so that in this case F' = F-
lac+). Such F
+ strains with F' are actually partial diploids or merozygotes. The use of F' elements to create partial diploids is called
sexduction or
F-duction. The origin of F' is shown in Figure 12.12, and may involve F' carrying upto one quarter of the bacterial chromosome. These merozygotes carrying F' have been used for recombination studies. F' elements are episomes and can be compared with plasmids discussed in
Plasmids, IS Elements, Transposons and Retroelements.

Fig. 12.12. Origin of F' factor (or F-lac) due to abnormal oullooping (a-c) and reintegration of F-lac+ into F-lac-strain.














