High resolution mapping
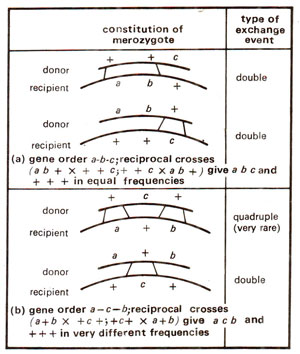
Fig. 12.16. Determination of gene order from relative frequencies of recombinants in reciprocal crosses (ab + x ++c).
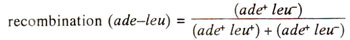
It has been shown that 1 minute on the map is equal to 20 per cent recombination, calculated as above.
If two genes are very close (b and c) with respect to a third gene (a), then it becomes difficult to determine the gene order (a—b—c or a—c—b), because the recombination frequencies between a and b and between a and c will be similar. This can be resolved by using reciprocal crosses : abc+ (Hfr) x a+b+c (F-) and a+b+c (Hfr) x abc+ (F-). In this case prototrophs will result in equal frequencies from two reciprocal crosses if the gene order is a-b-c and they will be produced in dramatically different frequencies, if the gene order is a-c-b (Fig. 12.16).




