Regulation of Methionine Biosynthesis
Methionine is a sulfur-containing essential amino acid, a building block of proteins
that also plays a fundamental role in many cellular processes. Through its
immediate catabolic product S-adenosyl methionine (SAM), methionine is a precursor
for the plant hormones ethylene and polyamines as well as for many
important secondary metabolites and vitamin B1. SAM is also a donor of a methyl
group to a number of cellular reactions, such as DNA methylation (Amir
et al.,
2002 and references therein). In plants, methionine can be converted into
S-methylmethionine (SMM), a metabolite that is believed to participate in sulfur
transport between sink and source tissues (Bourgis
et al., 1999), and also to control
the intracellular levels of SAM (Kocsis
et al., 2003; Ranocha
et al., 2001). Due to its
vital cellular importance, the methionine level is tightly regulated both by its
synthesis and catabolism. Methionine is an unstable amino acid with a very fast
half-life (Giovanelli
et al., 1985; Miyazaki and Yang, 1987).
Methionine receives its carbon and amino groups from
O-phosphohomoserine,
an intermediate metabolite in the aspartate family pathway, and its sulfur atom from cysteine (Fig. 3.2). These two skeleta are first combined by the enzyme
cystathionine γ-synthase (CGS) to form cystathionine. This is then converted by
cystathionine β-lyase into homocysteine, and converted by methionine synthase
into methionine, incorporating a methyl group from
N-methyltetrahydrofolate
(Fig. 3.2). Hence, the complex biosynthesis nature of methionine depends on
many regulatory metabolic steps, including the aspartate family pathway, cysteine
biosynthesis, and
N-methyltetrahydrofolate metabolism. Nevertheless, molecular
genetic and biochemical studies suggest that methionine biosynthesis is
regulated primarily by CGS as well as by a compound metabolic interaction
with threonine synthesis through a competition between CGS and threonine
synthase (TS) on their common substrate
O-phosphohomoserine (Fig. 3.2).
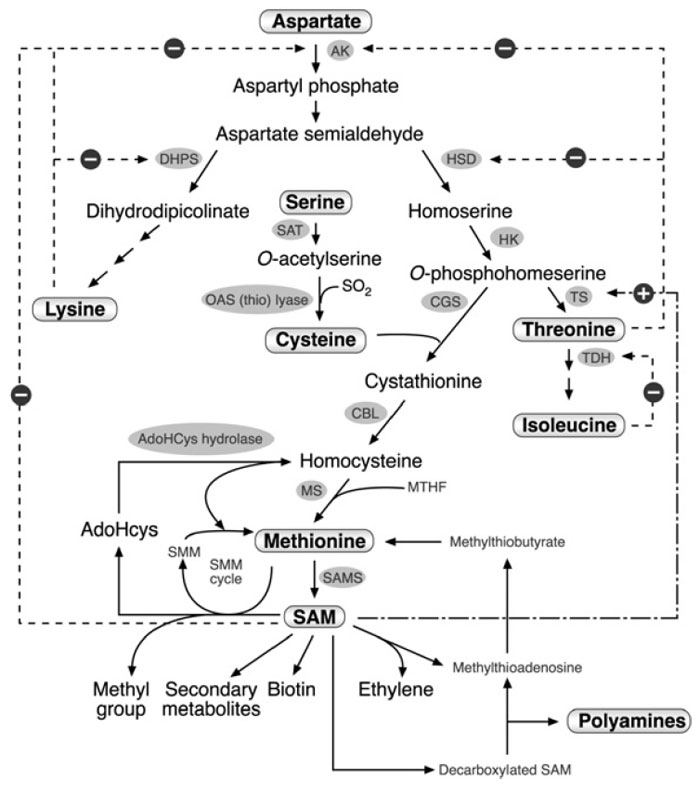 |
| FIGURE 3.2 Schematic diagram of the metabolic network containing the aspartate family
pathway, methionine metabolism, and last two steps in the cysteine biosynthesis. Only some of
the enzymes and metabolites are specified. Abbreviations: AK, aspartate kinase; DHPS,
dihydrodipicolinate synthase; HSD, homoserine dehydrogenase; HK, homoserine kinase; TS,
threonine synthase; TDH, threonine dehydratase; SAT, serine acetyl transferase; OAS (thio)
lyase; O-acetyl serine (thio) lyase; CGS, cystathionine γ-synthase; CBL, cystathionine β-lyase;
MS, methionine synthase, SAM, S-adenosyl methionine; SAMS, S-adenosyl methionine
synthase; AdoHcys, adenosylhomocysteine; SMM, S-methyl methionine; MTHF, methyltetrahydrofolate.
Dashed arrows with a ‘‘minus’’ sign represent feedback inhibition loops of key enzymes
in the network. The dashed and dotted arrow with the ‘‘plus’’ sign represents the stimulation of TS
activity by SAM. |





