Recombination in viruses
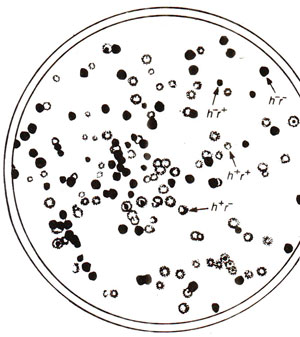
Fig. 12.23. Phage phenotypes produced by progeny of a cross h-r+ x h+r- (see text for details; redrawn from Stcnt, 1963)
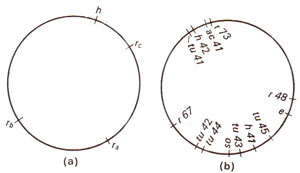
Fig. 12.24. (a) Circular genetic map of T2 phage, showing only four genes, (b) Circular genetic map of T4 phage; h = host range; ac = acridine resistance; tu = turbid plaques; os = resistance to osmotic shock; e = lysis defective.
The viruses, being very small particles, are often considered not suitable for inheritance studies due to non-availability of scorable traits. However, plaque morphology (large vs. small, fuzzy vs. sharp), host range and virulence are characteristic features of bacteriophages (plaque = clear area produced on opaque lawn of bacteria on the surface of a dish of solid medium), which have been used for inheritance and recombination studies. Inheritance and recombination in viruses can be illustrated by a cross in T2 phage, attempted by Alfred Hershey. This cross (h-r+ x h+r-) involved (i) host range (h+ can infect only strain 1 and h- can infect both strains 1 and 2); (ii) plaque morphology (r+ lyses slowly producing small plaques and r- lyses rapidly producing large plaques). The strain 1 was infected by both phage genotypes (mixed infection or double infection) and the lysate was analysed by spreading it onto a bacterial lawn composed of a mixture of strains 1 and 2 (h- will produce clear and h+ will produce cloudy plaques). Four plaque types were distinguishable (Fig. 12.23) :
(i) clear and small (h-r+), (ii) cloudy and large (h+r-), (iii) cloudy and small (h+r+) and (iv) clear and large (h-r-). The first two of these four are parental phenotypes and the last two are recombinants, so that the recombination frequency (RF) can be calculated as follows:

Fig. 12.23. Phage phenotypes produced by progeny of a cross h-r+ x h+r- (see text for details; redrawn from Stcnt, 1963)

Fig. 12.24. (a) Circular genetic map of T2 phage, showing only four genes, (b) Circular genetic map of T4 phage; h = host range; ac = acridine resistance; tu = turbid plaques; os = resistance to osmotic shock; e = lysis defective.
Recombination in T4 phage has also been studied by famous scientist S. Benzer, utilizing rll locus responsible for giving rise to rough morphology to the plaque (space cleared of bacteria due to phage infection). These studies enabled Benzer to work out the fine structure of gene at rll locus. A detailed discussion on this subject is presented in Fine Structure of Gene-at the Genetic Level (A New Concept of Allelomorphism).




