Phage - Displayed Antibody Libraries
Phage-displayed antibody libraries consist of large repertoires of Fab fragments (Barbas et al., 1991), single chain variable regions (scFv) (McCafferty et al., 1990), or diabodies (dimer scFv) (McGuinness et al., 1996) cloned into genetically engineered phage or phagemid vectors (Smith, 1985; Smith and Petrenko, 1997), and expressed on the surface of a bacteriophage. This phenotype-genotype linkage enables phage-displayed antibodies to be selected by multiple rounds of antigen-based affinity purification and amplification. In addition, phage-displayed libraries can be constructed bypassing the immune system and therefore can be targeted to self-antigens (Zeidel et al., 1995), as well as to nonimmunogenic and even toxic substances (Vaugham et al., 1996). Furthermore, being an in vitro technique, phage display technology makes it easier to build fully human antibody libraries (Holt et al., 2000). This article illustrates a general protocol for the generation of scFv libraries displayed on the surface of the pCGMT phage vector as part of either surface protein pIII (Gao et al., 1997; Mao et al., 1999; Gao et al., 1999) or pIX (Gao et al., 2002).
First-strand cDNA synthesis kit is from Amersham Pharmacia (Cat. No. 27-9261-01). Carbenicillin, tetracycline, kanamycin, and IPTG are from Research Products International (Cat. No. C46000-1.0, T17000/1.0, K22000-1.0, and I56000-5.0). Pfu DNA polymerase, Escherichia coli XL-1 blue, VCSM13 interferenceresistant helper phage, and total RNA purification kit are from Stratagene (Cat. No. 6000154, 200249, 200251, and 400790). Ultrapure agarose, electroporation cuvettes, 1-kb plus DNA ladder, T4 DNA ligase, bovine serum albumin (BSA), and 100 mM dNTP mix are from Invitrogen (Cat. No. 15510027, 15224017, P450-50, 10787018, 15561012, and 10216018). Dimethyl sulfoxide (DMSO) is from Aldrich (Cat. No. 27,043-1). SfiI is from New England Biolabs (Cat. No. R0123S). QIAquick gel extraction kit is from Qiagen (Cat. No. 28704). Tryptone is from Becton Dickinson (Cat. No. 211043). Yeast extract is from obtained EM Science (Cat. No. 1.03753.5007). Nonfat dry milk is from Bio- Rad (Cat. No. 170-6404). All the salts, glycerol, polyethylene glycol, Tween 20, ethanol, and glucose are from Sigma (Cat. No. G5516, P2139, P1379, 27,074-1, G5767). Petri dishes and Nunc MaxiSorb Immunotubes are from Fisher (Cat. No. 08-757-1000, and 12- 565-144). Polymerase chain reaction (PCR) tubes are from USA Scientific (Cat. No. 1402-4300). Nalgene 500-ml centrifuge bottles are from VWR (Cat. No. 21020-050). The thermocycler (Mastercycler Gradient) and benchtop centrifuge (5415C) are from Eppendorf. The electrophoresis chamber (Easy-Cast) is from Electrophoresis System. The electroporation apparatus (Gene Pulser II) is from Bio-Rad. The J2-H2 centrifuge is from Beckman.
A. Construction of a scFv Library
Solutions
- 2xYT: To make 1 liter, dissolve 16 g tryptone, 10 g yeast extract, and 5g NaCl in 900ml distilled water. After adjusting the pH to 7.0 with NaOH, bring the volume to 1 liter. Sterilize by autoclaving.
- SB medium: To make 1 liter, dissolve 30g tryptone, 20 g yeast extract, and 10g MOPS in 1 liter deionized water. Sterilize by autoclaving.
- LB-agar: To make 1 liter, dissolve 10 g tryptone, 5 g yeast, and 10 g NaCl in 900 ml distilled water. Add 15 g agar and adjust the pH to 7.0 with NaOH. Fill to 1 liter and autoclave. Allow to cool to a reasonable temperature and supplement with 1 ml carbenicillin stock, 10µg/ml tetracycline, and 2% glucose. Pour in petri dishes.
- SOC: To make 1 liter, dissolve 20g tryptone, 5g yeast, and 0.5 g NaCl in 900 ml distilled water. Add 10ml 250 mM KCl. Adjust the pH to 7.0, fill to 975 ml, and autoclave. Once cooled, add 5ml 2M MgCl2 and 20ml 1M glucose
- Phosphate-buffered saline (PBS): To make 1 liter, dissolve 1.44 g sodium phosphate, 0.24 g potassium phosphate, 0.2 g potassium chloride, and 8 g NaCl in 900ml distilled water. Adjust the pH to 7.4. Fill to 1 liter and autoclave.
- Blotto: To make 100 ml, dissolve 4 g nonfat dry milk in enough PBS to make 100ml final volume.
- 3 M NaOAc: Dissolve 24.61 g NaOAc in 90ml distilled water. Adjust the pH to 5.2. Fill to 100 ml and autoclave.
- 250 mM KCl: Dissolve 1.86 g KCl in 100 ml distilled water. Sterilize by autoclaving.
- 2M MgCl2: Dissolve 40.7g magnesium chloride hexahydrate in 100ml distilled water. Sterilize by autoclaving.
- 1M glucose: Dissolve 90g glucose in 500ml distilled water. Sterile filter.
- Carbenicillin stock: Dissolve 1 g carbenicillin in 10ml deionized water. Sterile filter. Store at -20°C.
- Tetracycline stock: Dissolve 50mg tetracycline in 10ml 70% ethanol. Store at-20°C.
- Kanamycin stock: Dissolve 500mg kanamycin in 10ml deionized water. Sterile filter. Store at -20°C.
- 0.5M IPTG stock: Dissolve 5 g isopropyl-β-Dthiogalactopyranoside (IPTG) in 42ml deionized water sterile filter. Store at -20°C.
- VCSM13 helper phage solution: Prepare this solution according to the vendor's instructions.
- Preparation of mRNA. Extract mRNA from either human peripheral blood lymphocytes (human library) or mouse spleen cells (murine library) using the RNA Purification Kit (Stratagene) according to the vendor's instructions. Prepare first-strand cDNA from the total RNA by using the First-strand cDNA Synthesis Kit (Pharmacia) and dT18 primer according to the manufacturer's recommendations.
- PCR amplification of antibody variable region genes (see Fig. 1). In a 250-µl PCR tube, combine 2µl of cDNA template, 2µl of one forward primer (100pmol/µl), 2µl of an equimolar mixture of the respective reverse primers (100pmol/µl), 200µM dNTPs, 5% DMSO, 10 µl 10× Pfu DNA polymerase buffer, and 5 units Pfu DNA polymerase (final volume: 100µl). For example, for the human heavy chain, set up 12 PCR reactions total, in which each HVH forward primer is combined with a mixture of HJH reverse primers. Set the temperature program as follows: denaturation at 94°C for 5 min; 30 cycles of amplification, including denaturation, 1 min, 94°C; annealing, 1 min, 50°C; extension, 1 min, 72°C and final extension at 72°C; for 10min. Run a 1% agarose gel and cut out the appropriate bands. Combine the heavy chain bands into one pool and the light chain bands (including Vλ and Vκ) into a separate pool. Purify using a Qiagen gel extraction kit.
- Construction of the scFv library (see Fig. 1). Assemble the scFv library by overlap PCR. In a 250-µl PCR tube, combine ~20 ng of each scFv fragment pool, 200µM dNTPs, 2µl 10× Pfu polymerase buffer, and 1 unit Pfu polymerase (final volume: 20µl). Set the temperature program as follows: denaturation at 94°C for 5 min; 5 cycles of amplification, including denaturation, 1 min, 94°C; annealing, 1 min; 55°C; and extension, 1.5min, 72°C. Add the outer primers HVH(Sfi) and HLJ(Sfi) [or MVH(Sfi) and MLJ(Sfi)] to a final concentration of 2 mM and bring the final volume to 50µl. Set the thermocycler for 30 more cycles of denaturation, 30s at 94°C; annealing, 30s at 60°C; extension, 1.5 min at 72°C; and final extension at 72°C for 10min. Gel purify the full-length scFv library on a 1% agarose gel using a Qiagen gel extraction kit.
- Digestion of the scFv library and vector. Digest both the scFv library and either pCGMT (pIII library; Gao et al., 1997) or pCGMT9 (pIX library; Gao et al., 2002) vector by combining 46 µl DNA, 6 µl 10x BSA, 6 µl 10x NE buffer 2, and 2 µl SfiI. Incubate the mixture for 2h or overnight at 50°C. Gel purify each digestion on a 1% agarose gel using a Qiagen gel extraction kit.
- Ligation into vector. Combine 5µl SfiI digested vector, 10 µl scFv library, 4 µl 5X ligase buffer, and 1 µl T4 DNA ligase for a final volume of 20µl. Incubate at 16°C for 2-10h. Add 2µl of 3M NaOAc, pH 5.2, and mix well. Add 40 µl absolute ethanol and store at -20°C for >20 min. Centrifuge at ≥12,000 rpm for 10 min. Decant supernatant and wash the pellet with 1 ml 70% ethanol, mixing well. Spin at≥12,000rpm for 1-2min and aspirate the solution. Air dry the pellet and resuspend in 10µl distilled water.
- Transformation into electrocompetent E. coli XL-1 Blue. Add 10µl of the ligation mixture to 500µl of a suspension of E. coli XL-1 Blue. Add 50-µl aliquots to 10 prechilled electroporation cuvettes. Electroporate at 2.5 kV per manufacture's instructions. Immediately add 200µl SOC medium to the cuvette and incubate at 37°C for 30min. Repeat as many times as necessary. Plate the cells on LB-agar plates supplemented with 2% glucose, 50 µg/ml carbenicillin, and 10 µg/ml tetracycline. Grow each plate overnight at 30°C.
- Library titer and storage. Scrape the clones off each plate with 10 ml SB medium supplemented with 10% glycerol and store at -70°C. Determine the size of the library by counting the number of independent transformants. To titer the library, serially dilute the library (1:10) in SB out to 10-9. Starting with the lowest dilution, add 100 µl onto a LB-carb plate and spread evenly. Incubate overnight at 37°C and count the colonyforming units for each dilution.
- Rescue of the scFv phage library. Inoculate ~5 × 105 cells oµained from the previously described glycerol stock into 100ml SB medium containing 2% glucose, 50µg/ml carbenicillin, and 10µg/ml tetracycline. Incubate the culture at 37°C on a reciprocal shaker until OD600 ~ 0.6 is reached. Add ~4 × 1013 plaqueforming units of VCSM13 helper phage and let incubate for 30min at room temperature and then for 90 min on the shaker at 37°C. Add 200 µl of 0.5 M IPTG and 140µl kanamycin and allow the culture to grow over-night at 30°C
- Phage precipitation. Pellet the cells by centrifuging at 7000rpm for 20min in a 400-ml centrifuge tube. Decant the supernatant into clean tubes. Dissolve 3g NaCl and 4 g PEG-8000 in each tube and place on ice for 30 min. Centrifuge at 7000 rpm for 20 min to pellet phage. Resuspend in an appropriate volume of PBS.
- Panning. Add 1 ml of ~5-50µg/ml antigen/ hapten to an immunotube and incubate overnight at 4°C. Remove the antigen solution and block the tube with 1 ml Blotto for 1 h at 37°C. Grow a culture of E. coli XL-1 Blue in 20 ml SB until the OD600 ~ 0.6. Save for phage titration and rescue. Remove the blocking solution and add 1012-1013 colony-forming units of phage library in PBS containing 1% nonfat dry milk and 3% BSA. Incubate at 37°C for 2h. Aspirate unbound phage solution and remove weakly bound phage by washing with 0.05% Tween 20 in PBS. Elute the tightly bound phage by either adding 1 ml of 0.1M glycine, pH 2.5, or adding a solution of free antigen. Incubate at room temperature for 10min. Remove the solution and neutralize with concentrated Tris base (usually 60µl 2M Tris, pH 8.0). Titrate the eluted phage by serially diluting (1:10) in SB out to 10-9. Starting with the lowest dilution, add 10µl diluted phage to 90µl E. coli XL-1 Blue. Plate 50µl onto a LB-carb plate and spread evenly. Incubate overnight at 37°C and count the colony-forming units for each dilution. Rescue the remaining eluted phage by adding to 20ml E. coli XL-1 Blue, mixing gently, and letting sit for 10 minutes at room temperature. Centrifuge at 3000rpm for 10 minutes and resuspend the pellet in approximately 500µ1 of the SB medium. Spread evenly on a LB-carb plate and incubate overnight at 37°C Rescue the library as outlined in step 7. This will be the input library for the next round of panning. Phage libraries are usually subjected to four to six rounds of panning. The number of rounds is dependent on enrichment as determined by library titer.
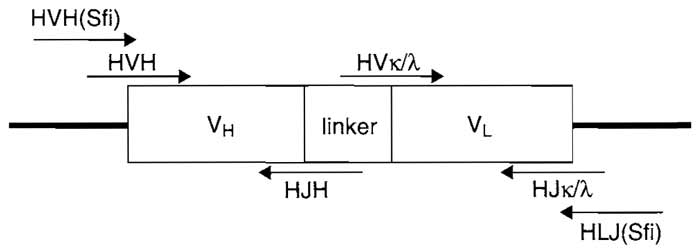 |
| FIGURE 1 scFv diagram indicating design and primer overlap. HVH, human variable heavy chain; HJH, human constant heavy chain; HVκ/λ, human variable light chain; HJκ/λ, human constant light chain; HVH(Sfi), human forward SfiI primer; HLJ(Sfi), human reverse SfiI primer. |
This procedure outlines the construction of a human or murine scFv library, including a list of suitable primers. The technique is also applicable to the formation of a Fab library with suitable primers (Barbas et al., 1991).
The stringency of the phage selection can be increased at the end of each round of panning in several ways. The first is to decrease the amount of antigen immobilized on the immunotubes. Similarly, the incubation time of the library with the antigen can be decreased. Alternatively, one can increase the number of washing steps and/or the amount of detergent in the wash buffer. Finally, one can use a progressively lower concentration of free antigen during the elution step. Note that when selecting for interactions influenced by the ionic strength of the medium, buffers other than PBS might be used for panning.
The number of transformations needed to obtain an appropriate library size has to be determined experimentally. Upon titration of the first transformation mixture, 30-50 colonies must be picked and the corresponding plasmids digested to determine the percentage of those containing inserts. Based on this number and the overall titer, one can determine the number of transformations needed to obtain ~109 independent transformants.
The quality of a library depends not only on the quantity of independent transformants, but also on diversity. In order to test such diversity, a set of 30-50 random clones should be sequenced so that the percentage of repeats can be determined. It is customary to define a library as satisfactory if it contains less than 10% repeated sequences.
- Take extreme care when working with RNA to prevent degradation by nucleases. This includes wearing gloves and using nuclease-free solutions.
- Failure in the primary PCR reaction is typically due to a low concentration of cDNA. Repeat the firststrand synthesis if necessary. It may also be necessary to adjust the annealing temperature. Note that lowering the annealing temperature allows for less fidelity in primer overlap.
- If overlap fails in step 3, check the concentrations of the various VH, Vκ, and Vλ fragments by running a small amount on a gel and examining the intensity of the bands. Adjust the amounts accordingly to give approximately equimolar concentrations.
- Scale the amount of phage preparation as necessary for library size.
References
Barbas, C. E, Kang, A. S., Lerner, R. A., and Benkovic, S. J. (1991). Assembly of combinatorial antibodies libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88, 7978-7982.
De Haard, H. J., van Neer, N., Reurs, A., Hufton, S. E., Roovers, R. C., Henderikx, P., de Bruine, A. P., Arends, J. W., and Hoogemboom, H. R. (1999). A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 274, 18218-18230.
Gao, C. S., Brummer, O., Mao, S. L., and Janda, K. D. (1999). Selection of human metalloantibodies from a combinatorial phage single-chain antibody library. J. Am. Chem. Soc. 121, 6517-6518.
Gao, C. S., Mao, S., Kaufmann, G., Wirsching, P., Lerner, R. A., and Janda, K. D. (2002). A method for the generation of combinatorial antibody libraries using pIX phage display. Proc. Natl. Acad. Sci. USA 99, 12612-12616.
Haidaris, C. G., Malone, J., Sherrill, L. A., Bliss, J. M., Gaspari, A. A., Insel, R. A., and Sullivan, M. A. (1999). Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J. lmmunol. Methods 257, 185-202.
Holt, L. J., Enever, C., de Wildt, R. M., and Tomlinson, I. M. (2000). The use of recombinant antibodies in proteomics. Curr. Opin. Biotech. 11,445-449.
Mao, S., Gao, C., Lo, C. H., Wirsching, P., Wong, C.-H., and Janda, K. D. (1999). Phage-display library selection of high-affinity human single-chain antibodies to tumor-associated carbohydrate antigens sialyl Lewis X and Lewis X. Proc. Natl. Acad. Sci. USA 96, 6953-6958.
Marks, J. D., Tristem, M., Karpas, A., and Winter, G. (1991). Oligonucleotides primers for polymerase chain reaction amplification of human immunoglobulin variable genes and design of familyspecific oligonucleotide probes. Eur. J. Immunol. 21,985-991.
McCafferty, J., Griffiths, A. D., Winter, G., and Chiswell, D. J. (1990). Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348, 552-554.
McGuinness, B. T., Walter, G., FitzGerald, K., Schuler, P., Mahoney, W., Duncan, A. R., and Hoogenboom, H. R. (1996). Phage diabody repertoires for selection of large numbers of bispecific antibody fragments. Nature Biotech. 14, 1149-1154.
Smith, G. P. (1985). Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315-1317.
Vaughan, T. J., Williams, A. J., Pritchard, K., Osbourn, J. K., Pope, A. R., Earnshaw, J. C., McCafferty, J., Hodits, R. A., Wilton, J., and Johnsons, K. S. (1996). Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nature Biotech. 14, 309-314.
Welschof, M., Terness, P., Kolbinger, E, Zewe, M., Dubel, S., Dorsam, H, Hain, C., Finger, M., Jung, M., Moldenhauer, G., Hayashi, N., Little, M., and Opelz, G. (1995). Amino acid sequence based PCR primers for amplification of rearranged human heavy and light chain immunoglobulin variable region genes. J. Immunol. Methods 179, 203-214.
Zeidel, M., Rey, E., Tami, J., Fischbach, M., and Sanz, I. (1995). Genetic and functional characterization of human autoantibodies using combinatorial phage display libraries. Ann. N. Y. Acad. Sci. 764, 559-564.




